Coordinating assembly of a bacterial macromolecular machine
- PMID: 18483484
- PMCID: PMC5963726
- DOI: 10.1038/nrmicro1887
Coordinating assembly of a bacterial macromolecular machine
Abstract
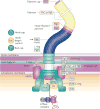
The assembly of large and complex organelles, such as the bacterial flagellum, poses the formidable problem of coupling temporal gene expression to specific stages of the organelle-assembly process. The discovery that levels of the bacterial flagellar regulatory protein FlgM are controlled by its secretion from the cell in response to the completion of an intermediate flagellar structure (the hook-basal body) was only the first of several discoveries of unique mechanisms that coordinate flagellar gene expression with assembly. In this Review, we discuss this mechanism, together with others that also coordinate gene regulation and flagellar assembly in Gram-negative bacteria.
Figures
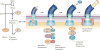
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

