A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels
- PMID: 18485064
- PMCID: PMC2574792
- DOI: 10.1111/j.1365-2958.2008.06289.x
A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels
Abstract
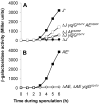
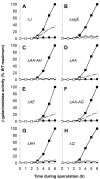
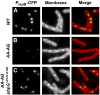
During spore formation in Bacillus subtilis, sigma(E)-directed gene expression in the mother-cell compartment of the sporangium triggers the activation of sigma(G) in the forespore by a pathway of intercellular signalling that is composed of multiple proteins of unknown function. Here, we confirm that the vegetative protein SpoIIIJ, the forespore protein SpoIIQ and eight membrane proteins (SpoIIIAA through SpoIIIAH) produced in the mother cell under the control of sigma(E) are ordinarily required for intercellular signalling. In contrast, an anti-sigma(G) factor previously implicated in the pathway is shown to be dispensable. We also present evidence suggesting that SpoIIIJ is a membrane protein translocase that facilitates the insertion of SpoIIIAE into the membrane. In addition, we report the isolation of a mutation that partially bypasses the requirement for SpoIIIJ and for SpoIIIAA through SpoIIIAG, but not for SpoIIIAH or SpoIIQ, in the activation of sigma(G). We therefore propose that under certain genetic conditions, SpoIIIAH and SpoIIQ can constitute a minimal pathway for the activation of sigma(G). Finally, based on the similarity of SpoIIIAH to a component of type III secretion systems, we speculate that signalling is mediated by a channel that links the mother cell to the forespore.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

