Aspects of coverage in medical DNA sequencing
- PMID: 18485222
- PMCID: PMC2430974
- DOI: 10.1186/1471-2105-9-239
Aspects of coverage in medical DNA sequencing
Abstract
Background: DNA sequencing is now emerging as an important component in biomedical studies of diseases like cancer. Short-read, highly parallel sequencing instruments are expected to be used heavily for such projects, but many design specifications have yet to be conclusively established. Perhaps the most fundamental of these is the redundancy required to detect sequence variations, which bears directly upon genomic coverage and the consequent resolving power for discerning somatic mutations.
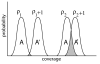
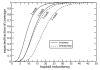
Results: We address the medical sequencing coverage problem via an extension of the standard mathematical theory of haploid coverage. The expected diploid multi-fold coverage, as well as its generalization for aneuploidy are derived and these expressions can be readily evaluated for any project. The resulting theory is used as a scaling law to calibrate performance to that of standard BAC sequencing at 8x to 10x redundancy, i.e. for expected coverages that exceed 99% of the unique sequence. A differential strategy is formalized for tumor/normal studies wherein tumor samples are sequenced more deeply than normal ones. In particular, both tumor alleles should be detected at least twice, while both normal alleles are detected at least once. Our theory predicts these requirements can be met for tumor and normal redundancies of approximately 26x and 21x, respectively. We explain why these values do not differ by a factor of 2, as might intuitively be expected. Future technology developments should prompt even deeper sequencing of tumors, but the 21x value for normal samples is essentially a constant.
Conclusion: Given the assumptions of standard coverage theory, our model gives pragmatic estimates for required redundancy. The differential strategy should be an efficient means of identifying potential somatic mutations for further study.
Figures


Similar articles
-
A general coverage theory for shotgun DNA sequencing.J Comput Biol. 2006 Jul-Aug;13(6):1177-96. doi: 10.1089/cmb.2006.13.1177. J Comput Biol. 2006. PMID: 16901236
-
A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal.PLoS Comput Biol. 2018 Feb 7;14(2):e1005965. doi: 10.1371/journal.pcbi.1005965. eCollection 2018 Feb. PLoS Comput Biol. 2018. PMID: 29415044 Free PMC article.
-
Whole-genome sequencing and variant discovery in C. elegans.Nat Methods. 2008 Feb;5(2):183-8. doi: 10.1038/nmeth.1179. Epub 2008 Jan 20. Nat Methods. 2008. PMID: 18204455
-
Genome sequencing-the dawn of a game-changing era.Heredity (Edinb). 2019 Jul;123(1):58-66. doi: 10.1038/s41437-019-0226-y. Epub 2019 Jun 12. Heredity (Edinb). 2019. PMID: 31189904 Free PMC article. Review.
-
The nematode Caenorhabditis elegans and its genome.Science. 1995 Oct 20;270(5235):410-4. doi: 10.1126/science.270.5235.410. Science. 1995. PMID: 7569995 Review.
Cited by
-
Genotype calling from next-generation sequencing data using haplotype information of reads.Bioinformatics. 2012 Apr 1;28(7):938-46. doi: 10.1093/bioinformatics/bts047. Epub 2012 Jan 27. Bioinformatics. 2012. PMID: 22285565 Free PMC article.
-
Effect of Next-Generation Exome Sequencing Depth for Discovery of Diagnostic Variants.Genomics Inform. 2015 Jun;13(2):31-9. doi: 10.5808/GI.2015.13.2.31. Epub 2015 Jun 30. Genomics Inform. 2015. PMID: 26175660 Free PMC article.
-
SomaticSniper: identification of somatic point mutations in whole genome sequencing data.Bioinformatics. 2012 Feb 1;28(3):311-7. doi: 10.1093/bioinformatics/btr665. Epub 2011 Dec 6. Bioinformatics. 2012. PMID: 22155872 Free PMC article.
-
Fast imputation using medium or low-coverage sequence data.BMC Genet. 2015 Jul 14;16:82. doi: 10.1186/s12863-015-0243-7. BMC Genet. 2015. PMID: 26168789 Free PMC article.
-
Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models.Open Biol. 2012 May;2(5):120061. doi: 10.1098/rsob.120061. Open Biol. 2012. PMID: 22724066 Free PMC article.
References
-
- Ley TJ, Minx PJ, Walter MJ, Ries RE, Sun H, McLellan M, DiPersio JF, Link DC, Tomasson MH, Graubert TA, McLeod H, Khoury H, Watson M, Shannon W, Trinkaus K, Heath S, Vardiman JW, Caligiuri MA, Bloomfield CD, Milbrandt JD, Mardis ER, Wilson RK. A Pilot Study of High-Throughput, Sequence-Based Mutational Profiling of Primary Human Acute Myeloid Leukemia Cell Genomes. Proceedings of the National Academy of Sciences. 2003;100:14275–14280. doi: 10.1073/pnas.2335924100. - DOI - PMC - PubMed
-
- Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, Busam D, Li K, Edwards JB, Eberhart C, Murphy KM, Tsiamouri A, Beeson K, Simpson AJG, Venter JC, Riggins GJ, Strausberg RL. Sequence Survey of Receptor Tyrosine Kinases Reveals Mutations in Glioblastomas. Proceedings of the National Academy of Sciences. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

