Molybdoproteomes and evolution of molybdenum utilization
- PMID: 18485362
- PMCID: PMC2670968
- DOI: 10.1016/j.jmb.2008.03.051
Molybdoproteomes and evolution of molybdenum utilization
Abstract
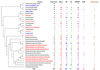
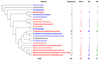
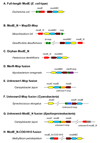
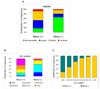
The trace element molybdenum (Mo) is utilized in many life forms, and it is a key component of several enzymes involved in nitrogen, sulfur, and carbon metabolism. With the exception of nitrogenase, Mo is bound in proteins to a pterin, thus forming the molybdenum cofactor (Moco) at the catalytic sites of molybdoenzymes. Although a number of molybdoenzymes are well characterized structurally and functionally, evolutionary analyses of Mo utilization are limited. Here, we carried out comparative genomic and phylogenetic analyses to examine the occurrence and evolution of Mo utilization in bacteria, archaea and eukaryotes at the level of (i) Mo transport and Moco utilization trait, and (ii) Mo-dependent enzymes. Our results revealed that most prokaryotes and all higher eukaryotes utilize Mo whereas many unicellular eukaryotes including parasites and most yeasts lost the ability to use this metal. In addition, eukaryotes have fewer molybdoenzyme families than prokaryotes. Dimethylsulfoxide reductase (DMSOR) and sulfite oxidase (SO) families were the most widespread molybdoenzymes in prokaryotes and eukaryotes, respectively. A distant group of the ModABC transport system, was predicted in the hyperthermophilic archaeon Pyrobaculum. ModE-type regulation of Mo uptake occurred in less than 30% of Moco-utilizing organisms. A link between Mo and selenocysteine utilization in prokaryotes was also identified wherein the selenocysteine trait was largely a subset of the Mo trait, presumably due to formate dehydrogenase, a Mo- and selenium-containing protein. Finally, analysis of environmental conditions and organisms that do or do not depend on Mo revealed that host-associated organisms and organisms with low G+C content tend to reduce their Mo utilization. Overall, our data provide new insights into Mo utilization and show its wide occurrence, yet limited use of this metal in individual organisms in all three domains of life.
Figures




References
-
- Enemark JH, Young CG. Bioinorganic chemistry of pterin-containing molybdenum and tungsten enzymes. Adv. Inorg. Chem. 1993;40:1–88.
-
- Hille R. The mononuclear molybdenum enzymes. Chem. Rev. 1996;96:2757–2816. - PubMed
-
- Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 2006;57:623–647. - PubMed
-
- Allen RM, Chatterjee R, Madden MS, Ludden PW, Shah VK. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Crit. Rev. Biotechnol. 1994;14:225–249. - PubMed
-
- Rajagopalan KV. Novel aspects of the biochemistry of the molybdenum cofactor. Adv. Enzym. 1991;64:215–290. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

