SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing
- PMID: 18485868
- PMCID: PMC2446403
- DOI: 10.1016/j.cell.2008.03.031
SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing
Abstract
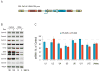
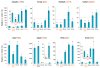
The survival of motor neurons (SMN) protein is essential for the biogenesis of small nuclear RNA (snRNA)-ribonucleoproteins (snRNPs), the major components of the pre-mRNA splicing machinery. Though it is ubiquitously expressed, SMN deficiency causes the motor neuron degenerative disease spinal muscular atrophy (SMA). We show here that SMN deficiency, similar to that which occurs in severe SMA, has unexpected cell type-specific effects on the repertoire of snRNAs and mRNAs. It alters the stoichiometry of snRNAs and causes widespread pre-mRNA splicing defects in numerous transcripts of diverse genes, preferentially those containing a large number of introns, in SMN-deficient mouse tissues. These findings reveal a key role for the SMN complex in RNA metabolism and in splicing regulation and indicate that SMA is a general splicing disease that is not restricted to motor neurons.
Figures




Comment in
-
Is good housekeeping the key to motor neuron survival?Cell. 2008 May 16;133(4):572-4. doi: 10.1016/j.cell.2008.05.002. Cell. 2008. PMID: 18485864
References
-
- Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J Biol Chem. 2002;277:31957–31962. - PubMed
-
- Balabanian S, Gendron NH, MacKenzie AE. Histologic and transcriptional assessment of a mild SMA model. Neurol Res. 2007;29:413–424. - PubMed
-
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol. 2006a;71:313–320. - PubMed
-
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006b;23:273–279. - PubMed
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

