The past and present of sodium energetics: may the sodium-motive force be with you
- PMID: 18485887
- PMCID: PMC2695506
- DOI: 10.1016/j.bbabio.2008.04.028
The past and present of sodium energetics: may the sodium-motive force be with you
Abstract
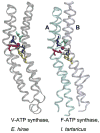
All living cells routinely expel Na(+) ions, maintaining lower concentration of Na(+) in the cytoplasm than in the surrounding milieu. In the vast majority of bacteria, as well as in mitochondria and chloroplasts, export of Na(+) occurs at the expense of the proton-motive force. Some bacteria, however, possess primary generators of the transmembrane electrochemical gradient of Na(+) (sodium-motive force). These primary Na(+) pumps have been traditionally seen as adaptations to high external pH or to high temperature. Subsequent studies revealed, however, the mechanisms for primary sodium pumping in a variety of non-extremophiles, such as marine bacteria and certain bacterial pathogens. Further, many alkaliphiles and hyperthermophiles were shown to rely on H(+), not Na(+), as the coupling ion. We review here the recent progress in understanding the role of sodium-motive force, including (i) the conclusion on evolutionary primacy of the sodium-motive force as energy intermediate, (ii) the mechanisms, evolutionary advantages and limitations of switching from Na(+) to H(+) as the coupling ion, and (iii) the possible reasons why certain pathogenic bacteria still rely on the sodium-motive force.
Figures
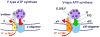
Similar articles
-
Evolutionary primacy of sodium bioenergetics.Biol Direct. 2008 Apr 1;3:13. doi: 10.1186/1745-6150-3-13. Biol Direct. 2008. PMID: 18380897 Free PMC article.
-
Animal plasma membrane energization by chemiosmotic H+ V-ATPases.J Exp Biol. 1997 Jan;200(Pt 2):203-16. doi: 10.1242/jeb.200.2.203. J Exp Biol. 1997. PMID: 9050228 Review.
-
Rotary Ion-Translocating ATPases/ATP Synthases: Diversity, Similarities, and Differences.Biochemistry (Mosc). 2020 Dec;85(12):1613-1630. doi: 10.1134/S0006297920120135. Biochemistry (Mosc). 2020. PMID: 33705299 Review.
-
Bacterial Na+ - or H+ -coupled ATP synthases operating at low electrochemical potential.Adv Microb Physiol. 2004;49:175-218. doi: 10.1016/S0065-2911(04)49004-3. Adv Microb Physiol. 2004. PMID: 15518831 Review.
-
The sodium cycle: a novel type of bacterial energetics.J Bioenerg Biomembr. 1989 Dec;21(6):635-47. doi: 10.1007/BF00762683. J Bioenerg Biomembr. 1989. PMID: 2687258 Review.
Cited by
-
Na+ riboswitches regulate genes for diverse physiological processes in bacteria.Nat Chem Biol. 2022 Aug;18(8):878-885. doi: 10.1038/s41589-022-01086-4. Epub 2022 Jul 25. Nat Chem Biol. 2022. PMID: 35879547 Free PMC article.
-
Na+-translocating membrane pyrophosphatases are widespread in the microbial world and evolutionarily precede H+-translocating pyrophosphatases.J Biol Chem. 2011 Jun 17;286(24):21633-42. doi: 10.1074/jbc.M111.244483. Epub 2011 Apr 28. J Biol Chem. 2011. PMID: 21527638 Free PMC article.
-
The role of energy in the emergence of biology from chemistry.Orig Life Evol Biosph. 2012 Oct;42(5):459-68. doi: 10.1007/s11084-012-9308-z. Orig Life Evol Biosph. 2012. PMID: 23100130 Free PMC article.
-
Extracellular electron transport-mediated Fe(III) reduction by a community of alkaliphilic bacteria that use flavins as electron shuttles.Appl Environ Microbiol. 2014 Jan;80(1):128-37. doi: 10.1128/AEM.02282-13. Epub 2013 Oct 18. Appl Environ Microbiol. 2014. PMID: 24141133 Free PMC article.
-
The Proton in Biochemistry: Impacts on Bioenergetics, Biophysical Chemistry, and Bioorganic Chemistry.Front Mol Biosci. 2021 Nov 26;8:764099. doi: 10.3389/fmolb.2021.764099. eCollection 2021. Front Mol Biosci. 2021. PMID: 34901158 Free PMC article. Review.
References
-
- Skulachev VP. Membrane Bioenergetics. Springer-Verlag; Berlin: 1988.
-
- Cramer WA, Knaff DB. Energy Transduction in Biological Membranes: A Textbook of Bioenergetics. Springer-Verlag; 1990.
-
- Skulachev VP. Membrane-linked energy transductions. Bioenergetic functions of sodium: H+ is not unique as a coupling ion. Eur J Biochem. 1985;151:199–208. - PubMed
-
- Skulachev VP. The sodium cycle: a novel type of bacterial energetics. J Bioenerg Biomembr. 1989;21:635–647. - PubMed
-
- Dimroth P. Bacterial energy transductions coupled to sodium ions. Res Microbiol. 1990;141:332–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

