cAMP-dependent signaling as a core component of the mammalian circadian pacemaker
- PMID: 18487196
- PMCID: PMC2735813
- DOI: 10.1126/science.1152506
cAMP-dependent signaling as a core component of the mammalian circadian pacemaker
Abstract
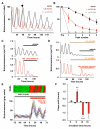
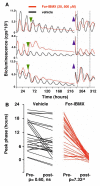
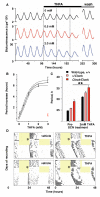
The mammalian circadian clockwork is modeled as transcriptional and posttranslational feedback loops, whereby circadian genes are periodically suppressed by their protein products. We show that adenosine 3',5'-monophosphate (cAMP) signaling constitutes an additional, bona fide component of the oscillatory network. cAMP signaling is rhythmic and sustains the transcriptional loop of the suprachiasmatic nucleus, determining canonical pacemaker properties of amplitude, phase, and period. This role is general and is evident in peripheral mammalian tissues and cell lines, which reveals an unanticipated point of circadian regulation in mammals qualitatively different from the existing transcriptional feedback model. We propose that daily activation of cAMP signaling, driven by the transcriptional oscillator, in turn sustains progression of transcriptional rhythms. In this way, clock output constitutes an input to subsequent cycles.
Figures

Comment in
-
Circadian rhythms. Integrating circadian timekeeping with cellular physiology.Science. 2008 May 16;320(5878):879-80. doi: 10.1126/science.1158619. Science. 2008. PMID: 18487177 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

