Exploring the suitability of coarse-grained techniques for the representation of protein dynamics
- PMID: 18487297
- PMCID: PMC2517050
- DOI: 10.1529/biophysj.107.119115
Exploring the suitability of coarse-grained techniques for the representation of protein dynamics
Abstract
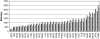
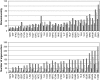
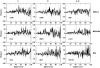
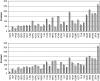
A systematic study of two coarse-grained techniques for the description of protein dynamics is presented. The two techniques exploit either Brownian or discrete molecular dynamics algorithms applied in the context of simple C(alpha)-C(alpha) potentials, like those used in coarse-grained normal mode analysis. Coarse-grained simulations of the flexibility of protein metafolds are compared to those computed with fully atomistic molecular dynamics simulations using state-of-the-art physical potentials and explicit solvent. Both coarse-grained models efficiently capture critical features of the protein dynamics.
Figures





References
-
- Daniel, R. M., R. V. Dumm, J. L. Finney, and C. J. Smith. 2003. The role of dynamics in enzyme activity. Annu. Rev. Biophys. Biomol. Struct. 32:69–92. - PubMed
-
- Eisenmesser, E. Z., D. A. Bosco, M. Akke, and D. Kern. 2002. Enzyme dynamics during catalysis. Science. 295:1520–1523. - PubMed
-
- Hinsen, K., A. Thomas, and M. J. Field. 1999. Analysis of domain motions in large proteins. Proteins. 34:369–382. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

