Identification of a cellobiose utilization gene cluster with cryptic beta-galactosidase activity in Vibrio fischeri
- PMID: 18487409
- PMCID: PMC2446528
- DOI: 10.1128/AEM.00190-08
Identification of a cellobiose utilization gene cluster with cryptic beta-galactosidase activity in Vibrio fischeri
Abstract
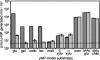
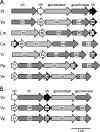
Cellobiose utilization is a variable trait that is often used to differentiate members of the family Vibrionaceae. We investigated how Vibrio fischeri ES114 utilizes cellobiose and found a cluster of genes required for growth on this beta-1,4-linked glucose disaccharide. This cluster includes genes annotated as a phosphotransferase system II (celA, celB, and celC), a glucokinase (celK), and a glucosidase (celG). Directly downstream of celCBGKA is celI, which encodes a LacI family regulator that represses cel transcription in the absence of cellobiose. When the celCBGKAI gene cluster was transferred to cellobiose-negative strains of Vibrio and Photobacterium, the cluster conferred the ability to utilize cellobiose. Genomic analyses of naturally cellobiose-positive Vibrio species revealed that V. salmonicida has a homolog of the celCBGKAI cluster, but V. vulnificus does not. Moreover, bioinformatic analyses revealed that CelG and CelK share the greatest homology with glucosidases and glucokinases in the phylum Firmicutes. These observations suggest that distinct genes for cellobiose utilization have been acquired by different lineages within the family Vibrionaceae. In addition, the loss of the celI regulator, but not the structural genes, attenuated the ability of V. fischeri to compete for colonization of its natural host, Euprymna scolopes, suggesting that repression of the cel gene cluster is important in this symbiosis. Finally, we show that the V. fischeri cellobioase (CelG) preferentially cleaves beta-d-glucose linkages but also cleaves beta-d-galactose-linked substrates such as 5-bromo-4-chloro-3-indolyl-beta-d-galactoside (X-gal), a finding that has important implications for the use of lacZ as a marker or reporter gene in V. fischeri.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ast, J. C., I. Cleenwerck, K. Engelbeen, H. Urbanczyk, F. L. Thompson, P. De Vos, and P. V. Dunlap. 2007. Photobacterium kishitanii sp. nov., a luminous marine bacterium symbiotic with deep-sea fishes. Int. J. Syst. Evol. Microbiol. 57:2073-2078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

