Prevention of amino acid conversion in SILAC experiments with embryonic stem cells
- PMID: 18487603
- PMCID: PMC2556023
- DOI: 10.1074/mcp.M800113-MCP200
Prevention of amino acid conversion in SILAC experiments with embryonic stem cells
Abstract
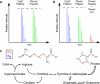
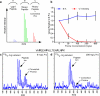
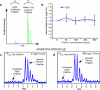
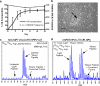
Recent studies using stable isotope labeling with amino acids in culture (SILAC) in quantitative proteomics have made mention of the problematic conversion of isotope-coded arginine to proline in cells. The resulting converted proline peptide divides the heavy peptide ion signal causing inaccuracy when compared with the light peptide ion signal. This is of particular concern as it can effect up to half of all peptides in a proteomic experiment. Strategies to both compensate for and limit the inadvertent conversion have been demonstrated, but none have been shown to prevent it. Additionally, these methods combined with SILAC labeling in general have proven problematic in their large scale application to sensitive cell types including embryonic stem cells (ESCs) from the mouse and human. Here, we show that by providing as little as 200 mg/liter L-proline in SILAC media, the conversion of arginine to proline can be rendered completely undetectable. At the same time, there was no compromise in labeling with isotope-coded arginine, indicating there is no observable back conversion from the proline supplement. As a result, when supplemented with proline, correct interpretation of "light" and "heavy" peptide ratios could be achieved even in the worst cases of conversion. By extending these principles to ESC culture protocols and reagents we were able to routinely SILAC label both mouse and human ESCs in the absence of feeder cells and without compromising the pluripotent phenotype. This study provides the simplest protocol to prevent proline artifacts in SILAC labeling experiments with arginine. Moreover, it presents a robust, feeder cell-free, protocol for performing SILAC experiments on ESCs from both the mouse and the human.
Figures

References
-
- Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 - PubMed
-
- Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 - PubMed
-
- Hwang, S. I., Lundgren, D. H., Mayya, V., Rezaul, K., Cowan, A. E., Eng, J. K., and Han, D. K. ( 2006) Systematic characterization of nuclear proteome during apoptosis: a quantitative proteomic study by differential extraction and stable isotope labeling. Mol. Cell Proteomics 5, 1131–1145 - PubMed
-
- Ong, S. E., Kratchmarova, I., and Mann, M. ( 2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 - PubMed
-
- Ong, S. E., and Mann, M. ( 2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

