Multidimensional fluorescence microscopy of multiple organelles in Arabidopsis seedlings
- PMID: 18489765
- PMCID: PMC2424051
- DOI: 10.1186/1746-4811-4-9
Multidimensional fluorescence microscopy of multiple organelles in Arabidopsis seedlings
Abstract
Background: The isolation of green fluorescent protein (GFP) and the development of spectral variants over the past decade have begun to reveal the dynamic nature of protein trafficking and organelle motility. In planta analyses of this dynamic process have typically been limited to only two organelles or proteins at a time in only a few cell types.
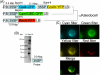
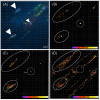
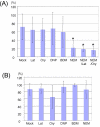
Results: We generated a transgenic Arabidopsis plant that contains four spectrally different fluorescent proteins. Nuclei, plastids, mitochondria and plasma membranes were genetically tagged with cyan, red, yellow and green fluorescent proteins, respectively. In addition, methods to track nuclei, mitochondria and chloroplasts and quantify the interaction between these organelles at a submicron resolution were developed. These analyzes revealed that N-ethylmaleimide disrupts nuclear-mitochondrial but not nuclear-plastids interactions in root epidermal cells of live Arabidopsis seedlings.
Conclusion: We developed a tool and associated methods for analyzing the complex dynamic of organelle-organelle interactions in real time in planta. Homozygous transgenic Arabidopsis (Kaleidocell) is available through Arabidopsis Biological Resource Center.
Figures




References
-
- Hiraoka Y, Swedlow JR, Paddy MR, Agard DA, Sedat JW. Three-dimensional multiple-wavelength fluorescence microscopy for the structural analysis of biological phenomena. Seminars in Cell Biology. 1991;2:153–165. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

