Cross-species de novo identification of cis-regulatory modules with GibbsModule: application to gene regulation in embryonic stem cells
- PMID: 18490265
- PMCID: PMC2493426
- DOI: 10.1101/gr.072769.107
Cross-species de novo identification of cis-regulatory modules with GibbsModule: application to gene regulation in embryonic stem cells
Abstract
We introduce the GibbsModule algorithm for de novo detection of cis-regulatory motifs and modules in eukaryote genomes. GibbsModule models the coexpressed genes within one species as sharing a core cis-regulatory motif and each homologous gene group as sharing a homologous cis-regulatory module (CRM), characterized by a similar composition of motifs. Without using a predetermined alignment result, GibbsModule iteratively updates the core motif shared by coexpressed genes and traces the homologous CRMs that contain the core motif. GibbsModule achieved substantial improvements in both precision and recall as compared with peer algorithms on a number of synthetic and real data sets. Applying GibbsModule to analyze the binding regions of the Krüppel-like factor (KLF) transcription factor in embryonic stem cells (ESCs), we discovered a motif that differs from a previously published KLF motif identified by a SELEX experiment, but the new motif is consistent with mutagenesis analysis. The SOX2 motif was found to be a collaborating motif to the KLF motif in ESCs. We used quantitative chromatin immunoprecipitation (ChIP) analysis to test whether GibbsModule could distinguish functional and nonfunctional binding sites. All seven tested binding sites in GibbsModule-predicted CRMs had higher ChIP signals as compared with the other seven tested binding sites located outside of predicted CRMs. GibbsModule is available at (http://biocomp.bioen.uiuc.edu/GibbsModule).
Figures


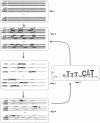
 (
( (score(n,n')) +
(score(n,n')) +  (score(n,n′))), where n, n′, and n″ are indicators of candidate CRMs in homologous sequences SeqA, orthA1, and orthA2, respectively. (X, O, #) Other motifs close to the core motif within a CRM. In Step 5, a new PSWM is calculated from the core motifs in the most conserved CRMs.
(score(n,n′))), where n, n′, and n″ are indicators of candidate CRMs in homologous sequences SeqA, orthA1, and orthA2, respectively. (X, O, #) Other motifs close to the core motif within a CRM. In Step 5, a new PSWM is calculated from the core motifs in the most conserved CRMs.
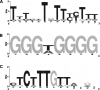
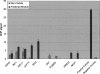
References
-
- Abeyta M.J., Clark A.T., Rodriguez R.T., Bodnar M.S., Pera R.A., Firpo M.T., Clark A.T., Rodriguez R.T., Bodnar M.S., Pera R.A., Firpo M.T., Rodriguez R.T., Bodnar M.S., Pera R.A., Firpo M.T., Bodnar M.S., Pera R.A., Firpo M.T., Pera R.A., Firpo M.T., Firpo M.T. Unique gene expression signatures of independently derived human embryonic stem cell lines. Hum. Mol. Genet. 2004;13:601–608. - PubMed
-
- Bailey T.L., Elkan C., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. - PubMed
-
- Banerji J., Rusconi S., Schaffner W., Rusconi S., Schaffner W., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. - PubMed
-
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Fry B., Meissner A., Wernig M., Plath K., Meissner A., Wernig M., Plath K., Wernig M., Plath K., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
