The complex genetics of multiple sclerosis: pitfalls and prospects
- PMID: 18490360
- PMCID: PMC2639203
- DOI: 10.1093/brain/awn081
The complex genetics of multiple sclerosis: pitfalls and prospects
Abstract
The genetics of complex disease is entering a new and exciting era. The exponentially growing knowledge and technological capabilities emerging from the human genome project have finally reached the point where relevant genes can be readily and affordably identified. As a result, the last 12 months has seen a virtual explosion in new knowledge with reports of unequivocal association to relevant genes appearing almost weekly. The impact of these new discoveries in Neuroscience is incalculable at this stage but potentially revolutionary. In this review, an attempt is made to illuminate some of the mysteries surrounding complex genetics. Although focused almost exclusively on multiple sclerosis all the points made are essentially generic and apply equally well, with relatively minor addendums, to any other complex trait, neurological or otherwise.
Figures
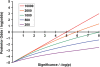
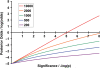
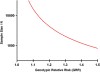


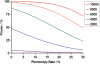


References
-
- Akesson E, Oturai A, Berg J, Fredrikson S, Andersen O, Harbo HF, et al. A genome-wide screen for linkage in Nordic sib-pairs with multiple sclerosis. Genes Immun. 2002;3:279–85. - PubMed
-
- Allcock RJ, Atrazhev AM, Beck S, de Jong PJ, Elliott JF, Forbes S, et al. The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens. 2002;59:520–1. - PubMed
-
- Ban M, Stewart GJ, Bennetts BH, Heard R, Simmons R, Maranian M, et al. A genome screen for linkage in Australian sibling-pairs with multiple sclerosis. Genes Immun. 2002;3:464–9. - PubMed
-
- Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–24. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

