Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale -a wild ancestor of cultivated buckwheat
- PMID: 18492277
- PMCID: PMC2430205
- DOI: 10.1186/1471-2229-8-59
Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale -a wild ancestor of cultivated buckwheat
Abstract
Background: Chloroplast genome sequences are extremely informative about species-interrelationships owing to its non-meiotic and often uniparental inheritance over generations. The subject of our study, Fagopyrum esculentum, is a member of the family Polygonaceae belonging to the order Caryophyllales. An uncertainty remains regarding the affinity of Caryophyllales and the asterids that could be due to undersampling of the taxa. With that background, having access to the complete chloroplast genome sequence for Fagopyrum becomes quite pertinent.
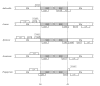
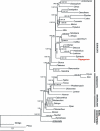
Results: We report the complete chloroplast genome sequence of a wild ancestor of cultivated buckwheat, Fagopyrum esculentum ssp. ancestrale. The sequence was rapidly determined using a previously described approach that utilized a PCR-based method and employed universal primers, designed on the scaffold of multiple sequence alignment of chloroplast genomes. The gene content and order in buckwheat chloroplast genome is similar to Spinacia oleracea. However, some unique structural differences exist: the presence of an intron in the rpl2 gene, a frameshift mutation in the rpl23 gene and extension of the inverted repeat region to include the ycf1 gene. Phylogenetic analysis of 61 protein-coding gene sequences from 44 complete plastid genomes provided strong support for the sister relationships of Caryophyllales (including Polygonaceae) to asterids. Further, our analysis also provided support for Amborella as sister to all other angiosperms, but interestingly, in the bayesian phylogeny inference based on first two codon positions Amborella united with Nymphaeales.
Conclusion: Comparative genomics analyses revealed that the Fagopyrum chloroplast genome harbors the characteristic gene content and organization as has been described for several other chloroplast genomes. However, it has some unique structural features distinct from previously reported complete chloroplast genome sequences. Phylogenetic analysis of the dataset, including this new sequence from non-core Caryophyllales supports the sister relationship between Caryophyllales and asterids.
Figures



References
-
- Soltis DE, Albert VA, Savolainen V, Hilu K, Qiu YL, Chase MW, Farris JS, Stefanoviæ S, Rice DW, Palmer JD, Soltis PS. Genome-scale data, angiosperm relationships, and 'ending incongruence': a cautionary tale in phylogenetics. Trends Plant Sci. 2004;9:477–483. doi: 10.1016/j.tplants.2004.08.008. - DOI - PubMed
-
- Degtjareva GV, Samigullin TH, Sokoloff DD, Valiejo-Roman CM. Gene sampling versus taxon sampling: is Amborella (Amborellaceae) a sister group to all other extant angiosperms? Bot Zhurn. 2004;89:896–907.
-
- Ohnishi O. Discovery of the wild ancestor of common buckwheat. Fagopyrum. 1991;11:5–10.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

