Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides
- PMID: 18492723
- PMCID: PMC2441798
- DOI: 10.1093/nar/gkn149
Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides
Abstract
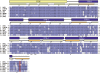
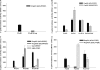
The transcriptional regulator PlcR and its cognate cell-cell signalling peptide PapR form a quorum-sensing system that controls the expression of extra-cellular virulence factors in various species of the Bacillus cereus group. PlcR and PapR alleles are clustered into four groups defining four pherotypes. However, the molecular basis for group specificity remains elusive, largely because the biologically relevant PapR form is not known. Here, we show that the in vivo active form of PapR is the C-terminal heptapeptide of the precursor, and not the pentapeptide, as previously suggested. Combining genetic complementation, anisotropy assays and structural analysis we provide a detailed functional and structural explanation for the group specificity of the PlcR-PapR quorum-sensing system. We further show that the C-terminal helix of the PlcR regulatory domain, specifically the 278 residue, in conjunction with the N-terminal residues of the PapR heptapeptide determines this system specificity. Variability in the specificity-encoding regions of plcR and papR genes suggests that selection and evolution of quorum-sensing systems play a major role in adaptation and ecology of Bacilli.
Figures



Similar articles
-
Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.Proc Natl Acad Sci U S A. 2013 Jan 15;110(3):1047-52. doi: 10.1073/pnas.1213770110. Epub 2012 Dec 31. Proc Natl Acad Sci U S A. 2013. PMID: 23277548 Free PMC article.
-
Specificity and polymorphism of the PlcR-PapR quorum-sensing system in the Bacillus cereus group.J Bacteriol. 2005 Feb;187(3):1182-7. doi: 10.1128/JB.187.3.1182-1187.2005. J Bacteriol. 2005. PMID: 15659693 Free PMC article.
-
The signaling peptide PapR is required for the activity of the quorum-sensor PlcRa in Bacillus thuringiensis.Microbiology (Reading). 2020 Apr;166(4):398-410. doi: 10.1099/mic.0.000883. Epub 2020 Jan 28. Microbiology (Reading). 2020. PMID: 32067627
-
Role of Sphingomyelinase in the Pathogenesis of Bacillus cereus Infection.Biol Pharm Bull. 2020;43(2):250-253. doi: 10.1248/bpb.b19-00762. Biol Pharm Bull. 2020. PMID: 32009113 Review.
-
What sets Bacillus anthracis apart from other Bacillus species?Annu Rev Microbiol. 2009;63:451-76. doi: 10.1146/annurev.micro.091208.073255. Annu Rev Microbiol. 2009. PMID: 19514852 Review.
Cited by
-
Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus.J Bacteriol. 2013 Apr;195(8):1845-55. doi: 10.1128/JB.02196-12. Epub 2013 Feb 8. J Bacteriol. 2013. PMID: 23396911 Free PMC article.
-
Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections.Sensors (Basel). 2013 Apr 18;13(4):5130-66. doi: 10.3390/s130405130. Sensors (Basel). 2013. PMID: 23598501 Free PMC article. Review.
-
Weak transcription of the cry1Ac gene in nonsporulating Bacillus thuringiensis cells.Appl Environ Microbiol. 2012 Sep;78(18):6466-74. doi: 10.1128/AEM.01229-12. Epub 2012 Jul 6. Appl Environ Microbiol. 2012. PMID: 22773626 Free PMC article.
-
How does Quorum Sensing of Intestinal Bacteria Affect Our Health and Mental Status?Microorganisms. 2022 Oct 5;10(10):1969. doi: 10.3390/microorganisms10101969. Microorganisms. 2022. PMID: 36296244 Free PMC article. Review.
-
Biosynthesis of peptide signals in gram-positive bacteria.Adv Appl Microbiol. 2010;71:91-112. doi: 10.1016/S0065-2164(10)71004-2. Epub 2010 Feb 20. Adv Appl Microbiol. 2010. PMID: 20378052 Free PMC article. Review.
References
-
- Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 1997;51:527–564. - PubMed
-
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. - PubMed
-
- Perego M, Brannigan JA. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides. 2001;22:1541–1547. - PubMed
-
- Granum PE, Lund T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997;157:223–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

