Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol
- PMID: 18492804
- PMCID: PMC2387273
- DOI: 10.1073/pnas.0802312105
Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol
Abstract
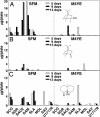
To identify the genes for biosynthesis of the off-flavor terpenoid alcohol, 2-methylisoborneol (2-MIB), the key genes encoding monoterpene cyclase were located in bacterial genome databases by using a combination of hidden Markov models, protein-family search, and the sequence alignment of their gene products. Predicted terpene cyclases were classified into three groups: sesquiterpene, diterpene, and other terpene cyclases. Genes of the terpene cyclase group that form an operon with a gene encoding S-adenosyl-l-methionine (SAM)-dependent methyltransferase were found in genome data of seven microorganisms belonging to actinomycetes, Streptomyces ambofaciens ISP5053, Streptomyces coelicolor A3(2), Streptomyces griseus IFO13350, Streptomyces lasaliensis NRRL3382R, Streptomyces scabies 87.22, Saccharopolyspora erythraea NRRL2338, and Micromonospora olivasterospora KY11048. Among six microorganisms tested, S. ambofaciens, S. coelicolor A3(2), S. griseus, and S. lasaliensis produced 2-MIB but M. olivasterospora produced 2-methylenebornane (2-MB) instead. The regions containing monoterpene cyclase and methyltransferase genes were amplified by PCR from S. ambofaciens, S. lasaliensis, and Saccharopolyspora erythraea, respectively, and their genes were heterologously expressed in Streptomyces avermitilis, which was naturally deficient of 2-MIB biosynthesis by insertion and deletion. All exoconjugants of S. avermitilis produced 2-MIB. Full-length recombinant proteins, monoterpene cyclase and methyltransferase of S. lasaliensis were expressed at high level in Escherichia coli. The recombinant methyltransferase catalyzed methylation at the C2 position of geranyl diphosphate (GPP) in the presence of SAM. 2-MIB was generated by incubation with GPP, SAM, recombinant methyltransferase, and terpene cyclase. We concluded that the biosynthetic pathway involves the methylation of GPP by GPP methyltransferase and its subsequent cyclization by monoterpene cyclase to 2-MIB.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


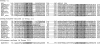


References
-
- Glasby JS. Encyclopedia of the Terpenoids. Chichester, U.K.: John Wiley; 1982.
-
- Sacchettini JC, Poulter CD. Biochemistry: Creating isoprenoid diversity. Science. 1997;277:1788–1789. - PubMed
-
- Schöller CEG, Gürtler H, Pedersen R, Molin S, Wilkins K. Volatile metabolites from actinomycetes. J Agric Food Chem. 2002;50:2615–2621. - PubMed
-
- Gerber NN. Volatile substances from actinomycetes. Crit Rev Microbiol. 1979;7:191–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

