FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron
- PMID: 18493046
- PMCID: PMC2390593
- DOI: 10.1534/genetics.107.083824
FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron
Abstract
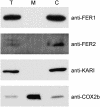
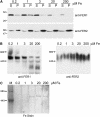
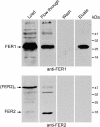
Two unlinked genes FER1 and FER2 encoding ferritin subunits were identified in the Chlamydomonas genome. An improved FER2 gene model, built on the basis of manual sequencing and incorporation of unplaced reads, indicated 49% identity between the ferritin subunits. Both FER1 and FER2 transcripts are increased in abundance as iron nutrition is decreased but the pattern for each gene is distinct. Using subunit-specific antibodies, we monitored expression at the protein level. In response to low iron, ferritin1 subunits and the ferritin1 complex are increased in parallel to the increase in FER1 mRNA. Nevertheless, the iron content of the ferritin1 complex is decreased. This suggests that increased expression results in increased capacity for iron binding in the chloroplast of iron-limited cells, which supports a role for ferritin1 as an iron buffer. On the other hand, ferritin2 abundance is decreased in iron-deprived cells, indicative of the operation of iron-nutrition-responsive regulation at the translational or post-translational level for FER2. Both ferritin subunits are plastid localized but ferritin1 is quantitatively recovered in soluble extracts of cells while ferritin2 is found in the particulate fraction. Partial purification of the ferritin1 complex indicates that the two ferritins are associated in distinct complexes and do not coassemble. The ratio of ferritin1 to ferritin2 is 70:1 in iron-replete cells, suggestive of a more dominant role of ferritin1 in iron homeostasis. The Volvox genome contains orthologs of each FER gene, indicating that the duplication of FER genes and potential diversification of function occurred prior to the divergence of species in the Volvocales.
Figures





Similar articles
-
Cloning, expression, and function of ferritins in the tick Haemaphysalis flava.Ticks Tick Borne Dis. 2022 Mar;13(2):101892. doi: 10.1016/j.ttbdis.2021.101892. Epub 2021 Dec 17. Ticks Tick Borne Dis. 2022. PMID: 34942560
-
Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii.Eukaryot Cell. 2002 Oct;1(5):736-57. doi: 10.1128/EC.1.5.736-757.2002. Eukaryot Cell. 2002. PMID: 12455693 Free PMC article.
-
Knockdown of proteins involved in iron metabolism limits tick reproduction and development.Proc Natl Acad Sci U S A. 2009 Jan 27;106(4):1033-8. doi: 10.1073/pnas.0807961106. Epub 2009 Jan 26. Proc Natl Acad Sci U S A. 2009. PMID: 19171899 Free PMC article.
-
Translational regulation of ferritin synthesis by iron.Enzyme. 1990;44(1-4):42-58. doi: 10.1159/000468746. Enzyme. 1990. PMID: 2133657 Review.
-
The ferritins: molecular properties, iron storage function and cellular regulation.Biochim Biophys Acta. 1996 Jul 31;1275(3):161-203. doi: 10.1016/0005-2728(96)00022-9. Biochim Biophys Acta. 1996. PMID: 8695634 Review.
Cited by
-
The expression of heterologous Fe (III) phytosiderophore transporter HvYS1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport.Plant Biotechnol J. 2017 Apr;15(4):423-432. doi: 10.1111/pbi.12637. Epub 2016 Oct 10. Plant Biotechnol J. 2017. PMID: 27633505 Free PMC article.
-
Iron uptake, transport and storage in marine brown algae.Biometals. 2023 Apr;36(2):371-383. doi: 10.1007/s10534-023-00489-7. Epub 2023 Mar 17. Biometals. 2023. PMID: 36930341 Review.
-
Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton.Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14652-7. doi: 10.1073/pnas.1506074112. Epub 2015 Nov 9. Proc Natl Acad Sci U S A. 2015. PMID: 26553998 Free PMC article.
-
The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii.Mol Cell Proteomics. 2013 Jan;12(1):65-86. doi: 10.1074/mcp.M112.021840. Epub 2012 Oct 13. Mol Cell Proteomics. 2013. PMID: 23065468 Free PMC article.
-
PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii.J Biol Chem. 2009 Nov 20;284(47):32770-81. doi: 10.1074/jbc.M109.050468. Epub 2009 Sep 25. J Biol Chem. 2009. PMID: 19783661 Free PMC article.
References
-
- Boughammoura, A., T. Franza, A. Dellagi, C. Roux, B. Matzanke-Markstein et al., 2007. Ferritins, bacterial virulence and plant defence. Biometals 20 347–353. - PubMed
-
- Boyd, P. W., T. Jickells, C. S. Law, S. Blain, E. A. Boyle et al., 2007. Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science 315 612–617. - PubMed
-
- Buchanan-Wollaston, V., and C. Ainsworth, 1997. Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridisation. Plant Mol. Biol. 33 821–834. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

