Oesophageal adenocarcinoma is associated with a deregulation in the MYC/MAX/MAD network
- PMID: 18493233
- PMCID: PMC2441969
- DOI: 10.1038/sj.bjc.6604398
Oesophageal adenocarcinoma is associated with a deregulation in the MYC/MAX/MAD network
Abstract
Oesophageal adenocarcinoma, which arises from an acquired columnar lesion, Barrett's metaplasia, is rising in incidence more rapidly than any other cancer in the Western world. Elevated expression of c-MYC has been demonstrated in oesophageal adenocarcinoma; however, the expression of other members of the MYC/MAX/MAD network has not been addressed. The aims of this work were to characterise the expression of c-MYC, MAX and the MAD family in adenocarcinoma development and assess the effects of overexpression on cellular behaviour. mRNA expression in samples of Barrett's metaplasia and oesophageal adenocarcinoma were examined by qRT-PCR. Semi-quantitative immunohistochemistry and western blotting were used to examine cellular localisation and protein levels. Cellular proliferation and mRNA expression were determined in SEG1 cells overexpressing c-MYCER or MAD1 using a bromodeoxyuridine assay and qRT-PCR, respectively. Consistent with previous work expression of c-MYC was deregulated in oesophageal adenocarcinoma. Paradoxically, increased expression of putative c-MYC antagonists MAD1 and MXI1 was observed in tumour specimens. Overexpression of c-MYC and MAD proteins in SEG1 cells resulted in differential expression of MYC/MAX/MAD network members and reciprocal changes in proliferation. In conclusion, the expression patterns of c-MYC, MAX and the MAD family were shown to be deregulated in the oesophageal cancer model.
Figures
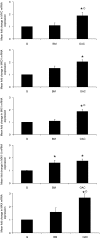


 ) or MXD1 (
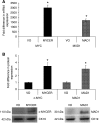
) or MXD1 ( ) mRNA expression. (B) Western blotting demonstrated the expression the chimeric protein in SEG1-MYCER or MAD1 in SEG1-MAD1. Densitometric scanning approximated the fold increase in expression; a representative blot is also shown. Values represent the mean of two experiments each performed in triplicate ±1 s.e.m. * denotes statistical significance (P<0.05).
) mRNA expression. (B) Western blotting demonstrated the expression the chimeric protein in SEG1-MYCER or MAD1 in SEG1-MAD1. Densitometric scanning approximated the fold increase in expression; a representative blot is also shown. Values represent the mean of two experiments each performed in triplicate ±1 s.e.m. * denotes statistical significance (P<0.05).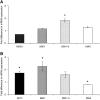
 , MXI1
, MXI1
 , MXI-0
, MXI-0
 and MAX
and MAX
 mRNA in SEG1 cells transiently overexpressing MYCER. Relative gene expression is expressed as a ratio of SEG1-MYCER not stimulated using 4OHT normalised to one. (B) Expression of MYC
mRNA in SEG1 cells transiently overexpressing MYCER. Relative gene expression is expressed as a ratio of SEG1-MYCER not stimulated using 4OHT normalised to one. (B) Expression of MYC
 , MXI1
, MXI1
 , MXI-0
, MXI-0
 and MAX
and MAX
 mRNA was assessed in SEG1 cells transiently overexpressing MAD1. Relative gene expression is expressed as a ratio of mock transfected cells normalised to one. Data represent the mean of two independent experiments each performed in triplicate ±1 s.e.m. * denotes statistical significance (P<0.05).
mRNA was assessed in SEG1 cells transiently overexpressing MAD1. Relative gene expression is expressed as a ratio of mock transfected cells normalised to one. Data represent the mean of two independent experiments each performed in triplicate ±1 s.e.m. * denotes statistical significance (P<0.05).References
-
- Ayer DE, Kretzner L, Eisenman RN (1993) Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72: 211–222 - PubMed
-
- Bartsch D, Peiffer SL, Kaleem Z, Wells Jr SA, Goodfellow PJ (1996) Mxi1 tumor suppressor gene is not mutated in primary pancreatic adenocarcinoma. Cancer Lett 102: 73–76 - PubMed
-
- Benson LQ, Coon MR, Krueger LM, Han GC, Sarnaik AA, Wechsler DS (1999) Expression of MXI1, a Myc antagonist, is regulated by Sp1 and AP2. J Biol Chem 274: 28794–28802 - PubMed
-
- Berberich S, Hyde-DeRuyscher N, Espenshade P, Cole M (1992) Max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene 7: 775–779 - PubMed
-
- Bernasconi NL, Wormhoudt TA, Laird-Offringa IA (2000) Post-transcriptional deregulation of myc genes in lung cancer cell lines. Am J Respir Cell Mol Biol 23: 560–565 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

