Biochemical visualization of cell surface molecular clustering in living cells
- PMID: 18495923
- PMCID: PMC2396715
- DOI: 10.1073/pnas.0710346105
Biochemical visualization of cell surface molecular clustering in living cells
Abstract
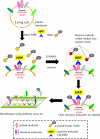
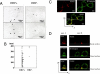
Many plasma membrane-resident molecules cluster with other molecules to collaborate in a variety of biological events. We herein report a sensitive and simple method to identify components of cell surface molecular clusters in living cells. This method includes a recently established reaction, called the enzyme-mediated activation of radical source (EMARS), to label molecules within a limited distance ( approximately 200-300 nm) from the probed molecule on which HRP is set. Because the size of this active area is close to that of the reported membrane clusters, it is suggested that the labeled molecules cluster with the probed molecule in the same membrane domain. A combination of the EMARS reaction and antibody array analysis demonstrated that many kinds of receptor tyrosine kinases (RTKs) formed clusters with beta1 integrin in HeLa S3 cells. A similar antibody array analysis after the EMARS reaction with three HRP-labeled antibodies against growth factor receptors showed the patterns of biotinylated RTKs to be substantially different from each other. These results suggest that different types of cell surface molecular clusters can thus be distinguished using the EMARS reaction. Therefore, the present "biochemical visualization" method is expected to be a powerful tool to elucidate molecular clustering on the cell surface of living cells in various contexts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. - PubMed
-
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. - PubMed
-
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

