Multiple functions of DNA polymerases
- PMID: 18496613
- PMCID: PMC2391090
- DOI: 10.1080/07352680701252817
Multiple functions of DNA polymerases
Abstract
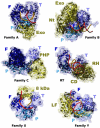
The primary role of DNA polymerases is to accurately and efficiently replicate the genome in order to ensure the maintenance of the genetic information and its faithful transmission through generations. This is not a simple task considering the size of the genome and its constant exposure to endogenous and environmental DNA damaging agents. Thus, a number of DNA repair pathways operate in cells to protect the integrity of the genome. In addition to their role in replication, DNA polymerases play a central role in most of these pathways. Given the multitude and the complexity of DNA transactions that depend on DNA polymerase activity, it is not surprising that cells in all organisms contain multiple highly specialized DNA polymerases, the majority of which have only recently been discovered. Five DNA polymerases are now recognized in Escherichia coli, 8 in Saccharomyces cerevisiae, and at least 15 in humans. While polymerases in bacteria, yeast and mammalian cells have been extensively studied much less is known about their counterparts in plants. For example, the plant model organism Arabidopsis thaliana is thought to contain 12 DNA polymerases, whose functions are mostly unknown. Here we review the properties and functions of DNA polymerases focusing on yeast and mammalian cells but paying special attention to the plant enzymes and the special circumstances of replication and repair in plant cells.
Figures

References
-
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2006;13:35–43. - PubMed
-
- Autexier C, Lue NF. The Structure and Function of Telomerase Reverse Transcriptase. Annu. Rev. Biochem. 2006 - PubMed
-
- Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
