Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins
- PMID: 18497292
- PMCID: PMC2754768
- DOI: 10.1126/science.1158265
Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins
Abstract
Nitric oxide acts substantially in cellular signal transduction through stimulus-coupled S-nitrosylation of cysteine residues. The mechanisms that might subserve protein denitrosylation in cellular signaling remain uncharacterized. Our search for denitrosylase activities focused on caspase-3, an exemplar of stimulus-dependent denitrosylation, and identified thioredoxin and thioredoxin reductase in a biochemical screen. In resting human lymphocytes, thioredoxin-1 actively denitrosylated cytosolic caspase-3 and thereby maintained a low steady-state amount of S-nitrosylation. Upon stimulation of Fas, thioredoxin-2 mediated denitrosylation of mitochondria-associated caspase-3, a process required for caspase-3 activation, and promoted apoptosis. Inhibition of thioredoxin-thioredoxin reductases enabled identification of additional substrates subject to endogenous S-nitrosylation. Thus, specific enzymatic mechanisms may regulate basal and stimulus-induced denitrosylation in mammalian cells.
Figures
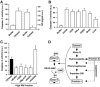
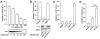


Comment in
-
Biochemistry. SNO removal.Science. 2008 May 23;320(5879):1019-20. doi: 10.1126/science.1159246. Science. 2008. PMID: 18497281 No abstract available.
References
-
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat Rev Mol Cell Biol. 2005;6:150. - PubMed
-
- Mannick JB, et al. Science. 1999;284:651. - PubMed
-
- Erwin PA, Lin AJ, Golan DE, Michel T. J Biol Chem. 2005;280:19888. - PubMed
-
- Hoffmann J, Haendeler J, Zeiher AM, Dimmeler S. J Biol Chem. 2001;276:41383. - PubMed
-
- Kim JE, Tannenbaum SR. J Biol Chem. 2004;279:9758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

