Roles of histidines 154 and 189 and aspartate 139 in the active site of serine acetyltransferase from Haemophilus influenzae
- PMID: 18498176
- PMCID: PMC2854626
- DOI: 10.1021/bi800075c
Roles of histidines 154 and 189 and aspartate 139 in the active site of serine acetyltransferase from Haemophilus influenzae
Abstract
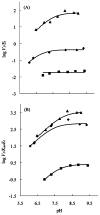
A crystal structure of serine acetyltransferase (SAT) with cysteine bound in the serine subsite of the active site shows that both H154 and H189 are within hydrogen-bonding distance to the cysteine thiol [Olsen, L. R., Huang, B., Vetting, M. W., and Roderick, S. L. (2004) Biochemistry 43, 6013 -6019]. In addition, H154 is in an apparent dyad linkage with D139. The structure suggests that H154 is the most likely catalytic general base and that H189 and D139 may also play important roles during the catalytic reaction. Site-directed mutagenesis was performed to mutate each of these three residues to Asn, one at a time. The V1/Et value of all of the single mutant enzymes decreased, with the largest decrease (approximately 1240-fold) exhibited by the H154N mutant enzyme. Mutation of both histidines, H154N/H189N, gave a V1/Et approximately 23700-fold lower than that of the wild-type enzyme. An increase in K Ser was observed for the H189N, D139N, and H154N/H189N mutant enzymes, while the H154N mutant enzyme gave an 8-fold decrease in K Ser. For all three single mutant enzymes, V1/Et and V1/K Ser Et decrease at low pH and give a pKa of about 7, while the V1/Et of the double mutant enzyme was pH independent. The solvent deuterium kinetic isotope effects on V 1 and V1/K Ser decreased compared to wild type for the H154N mutant enzyme and increased for the H189N mutant enzyme but was about the same as that of wild type for D139N and H154N/H189N. Data suggest that H154, H189, and D139 play different catalytic roles for SAT. H154 likely serves as a general base, accepting a proton from the beta-hydroxyl of serine as the tetrahedral intermediate is formed upon nucleophilic attack on the thioester carbonyl of acetyl-CoA. However, activity is not completely lost upon elimination of H154, and thus, H189 may be able to serve as a backup general base at a lower efficiency compared to H154; it also aids in binding and orienting the serine substrate. Aspartate 139, in dyad linkage with H154, likely facilitates catalysis by increasing the basicity of H154.
Figures
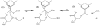
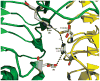

References
-
- Kredich NM, Tomkins GM. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–4965. - PubMed
-
- Smith IK, Thompson JF. Purification and characterization of L-serine transacetylase and O-acetyl-L-serine sulfhydrylase from kidney bean seedlings (Phaseolus vulgaris) Biochim Biophys Acta. 1971;227:288–295. - PubMed
-
- Vaara M. Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol Lett. 1992;76:249–254. - PubMed
-
- Raetz CR, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

