Multisite phosphorylation regulates Bim stability and apoptotic activity
- PMID: 18498746
- PMCID: PMC2453504
- DOI: 10.1016/j.molcel.2008.03.025
Multisite phosphorylation regulates Bim stability and apoptotic activity
Abstract
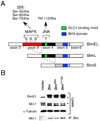
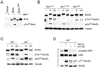
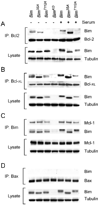
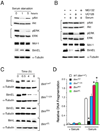
The proapoptotic BH3-only protein Bim is established to be an important mediator of signaling pathways that induce cell death. Multisite phosphorylation of Bim by several members of the MAP kinase group is implicated as a regulatory mechanism that controls the apoptotic activity of Bim. To test the role of Bim phosphorylation in vivo, we constructed mice with a series of mutant alleles that express phosphorylation-defective Bim proteins. We show that mutation of the phosphorylation site Thr-112 causes decreased binding of Bim to the antiapoptotic protein Bcl2 and can increase cell survival. In contrast, mutation of the phosphorylation sites Ser-55, Ser-65, and Ser-73 can cause increased apoptosis because of reduced proteasomal degradation of Bim. Together, these data indicate that phosphorylation can regulate Bim by multiple mechanisms and that the phosphorylation of Bim on different sites can contribute to the sensitivity of cellular apoptotic responses.
Figures


References
-
- Bouillet P, Cory S, Zhang LC, Strasser A, Adams JM. Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Dev Cell. 2001a;1:645–653. - PubMed
-
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. - PubMed
-
- Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

