Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1
- PMID: 18499658
- PMCID: PMC2475708
- DOI: 10.1074/jbc.M802696200
Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1
Abstract
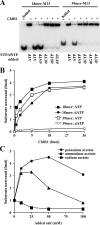
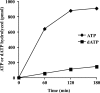
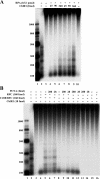
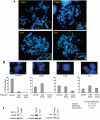
Human ChlR1 (hChlR1), a member of the DEAD/DEAH subfamily of helicases, was shown to interact with components of the cohesin complex and play a role in sister chromatid cohesion. In order to study the biochemical and biological properties of hChlR1, we purified the protein from 293 cells and demonstrated that hChlR1 possesses DNA-dependent ATPase and helicase activities. This helicase translocates on single-stranded DNA in the 5' to 3' direction in the presence of ATP and, to a lesser extent, dATP. Its unwinding activity requires a 5'-singlestranded region for helicase loading, since flush-ended duplex structures do not support unwinding. The helicase activity of hChlR1 is capable of displacing duplex regions up to 100 bp, which can be extended to 500 bp by RPA or the cohesion establishment factor, the Ctf18-RFC (replication factor C) complex. We show that hChlR1 interacts with the hCtf18-RFC complex, human proliferating cell nuclear antigen, and hFen1. The interactions between Fen1 and hChlR1 stimulate the flap endonuclease activity of Fen1. Selective depletion of either hChlR1 or Fen1 by targeted small interfering RNA treatment results in the precocious separation of sister chromatids. These findings are consistent with a role of hChlR1 in the establishment of sister chromatid cohesion and suggest that its action may contribute to lagging strand processing events important in cohesion.
Figures


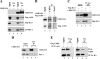
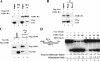
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

