Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis
- PMID: 18499678
- PMCID: PMC2459274
- DOI: 10.1074/jbc.M802940200
Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis
Abstract
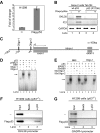
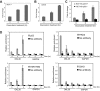
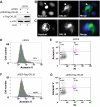
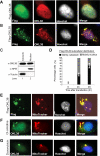
Protein Arg methyltransferases function as coactivators of the tumor suppressor p53 to regulate gene expression. Peptidylarginine deiminase 4 (PAD4/PADI4) counteracts the functions of protein Arg methyltransferases in gene regulation by deimination and demethylimination. Here we show that the expression of a tumor suppressor gene, OKL38, is activated by the inhibition of PAD4 or the activation of p53 following DNA damage. Chromatin immunoprecipitation assays showed a dynamic change of p53 and PAD4 occupancy and histone Arg modifications at the OKL38 promoter during DNA damage, suggesting a direct role of PAD4 and p53 in the expression of OKL38. Furthermore, we found that OKL38 induces apoptosis through localization to mitochondria and induction of cytochrome c release. Together, our studies identify OKL38 as a novel p53 target gene that is regulated by PAD4 and plays a role in apoptosis.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

