Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype
- PMID: 18500333
- PMCID: PMC2613942
- DOI: 10.1038/nature06973
Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype
Abstract
Understanding the molecular underpinnings of cancer is of critical importance to the development of targeted intervention strategies. Identification of such targets, however, is notoriously difficult and unpredictable. Malignant cell transformation requires the cooperation of a few oncogenic mutations that cause substantial reorganization of many cell features and induce complex changes in gene expression patterns. Genes critical to this multifaceted cellular phenotype have therefore only been identified after signalling pathway analysis or on an ad hoc basis. Our observations that cell transformation by cooperating oncogenic lesions depends on synergistic modulation of downstream signalling circuitry suggest that malignant transformation is a highly cooperative process, involving synergy at multiple levels of regulation, including gene expression. Here we show that a large proportion of genes controlled synergistically by loss-of-function p53 and Ras activation are critical to the malignant state of murine and human colon cells. Notably, 14 out of 24 'cooperation response genes' were found to contribute to tumour formation in gene perturbation experiments. In contrast, only 1 in 14 perturbations of the genes responding in a non-synergistic manner had a similar effect. Synergistic control of gene expression by oncogenic mutations thus emerges as an underlying key to malignancy, and provides an attractive rationale for identifying intervention targets in gene networks downstream of oncogenic gain- and loss-of-function mutations.
Figures
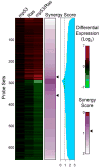
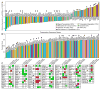


Comment in
-
Cancer: Deconstructing oncogenesis.Nature. 2008 Jun 19;453(7198):995-6. doi: 10.1038/453995a. Nature. 2008. PMID: 18563141 No abstract available.
References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

