Mouse segmental duplication and copy number variation
- PMID: 18500340
- PMCID: PMC2574762
- DOI: 10.1038/ng.172
Mouse segmental duplication and copy number variation
Abstract
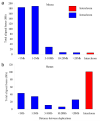
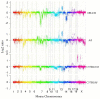
Detailed analyses of the clone-based genome assembly reveal that the recent duplication content of mouse (4.94%) is now comparable to that of human (5.5%), in contrast to previous estimates from the whole-genome shotgun sequence assembly. However, the architecture of mouse and human genomes differs markedly: most mouse duplications are organized into discrete clusters of tandem duplications that show depletion of genes and transcripts and enrichment of long interspersed nuclear element (LINE) and long terminal repeat (LTR) retroposons. We assessed copy number variation of the C57BL/6J duplicated regions within 15 mouse strains previously used for genetic association studies, sequencing and the Mouse Phenome Project. We determined that over 60% of these base pairs are polymorphic among the strains (on average, there was 20 Mb of copy-number-variable DNA between different mouse strains). Our data suggest that different mouse strains show comparable, if not greater, copy number polymorphism when compared to human; however, such variation is more locally restricted. We show large and complex patterns of interstrain copy number variation restricted to large gene families associated with spermatogenesis, pregnancy, viviparity, pheromone signaling and immune response.
Figures








References
-
- Bailey JA, Eichler EE. Genome-wide detection of segmental duplication within mammalian organisms. In: Ebert J, editor. Proceedings of the 68th Cold Spring Harbor Symposium: Genome of Homo sapiens; New York: Cold Spring Harbor Press; 2003. - PubMed
-
- She X, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–30. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

