Endogenous RNA interference provides a somatic defense against Drosophila transposons
- PMID: 18501606
- PMCID: PMC2812477
- DOI: 10.1016/j.cub.2008.05.006
Endogenous RNA interference provides a somatic defense against Drosophila transposons
Abstract
Background: Because of the mutagenic consequences of mobile genetic elements, elaborate defenses have evolved to restrict their activity. A major system that controls the activity of transposable elements (TEs) in flies and vertebrates is mediated by Piwi-interacting RNAs (piRNAs), which are approximately 24-30 nucleotide RNAs that are bound by Piwi-class effectors. The piRNA system is thought to provide primarily a germline defense against TE activity.
Results: Here, we describe a second system that represses Drosophila TEs by using endogenous small interfering RNAs (siRNAs), which are 21 nucleotide, 3'-end-modified RNAs that are dependent on Dicer-2 and Argonaute-2. In contrast to piRNAs, we find that the TE-siRNA system is active in somatic tissues, and particularly so in various immortalized cell lines. Analysis of the patterns and properties of TE-derived small RNAs reveals further distinctions between TE regions and genomic loci that are converted into piRNAs and siRNAs, respectively. Finally, functional tests show that many transposon transcripts accumulate to higher levels in cells and animal tissues that are deficient for Dicer-2 or Argonaute-2.
Conclusions: Drosophila utilizes two small-RNA systems to restrict transposon activity in the germline (mostly via piRNAs) and in the soma (mostly via siRNAs).
Figures
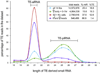
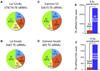
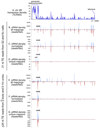
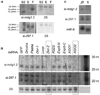

Comment in
-
RNA interference: endogenous siRNAs derived from transposable elements.Curr Biol. 2008 Jul 8;18(13):R561-3. doi: 10.1016/j.cub.2008.05.035. Curr Biol. 2008. PMID: 18606126
References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. - PubMed
-
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA Silencing Pathways. Cell. 2004;117:69–81. - PubMed
-
- Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

