Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G
- PMID: 18501607
- PMCID: PMC2491687
- DOI: 10.1016/j.cub.2008.04.073
Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G
Abstract
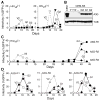
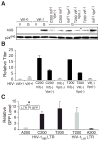
The human APOBEC3G protein restricts the replication of Vif-deficient HIV-1 by deaminating nascent viral cDNA cytosines to uracils, leading to viral genomic strand G-to-A hypermutations. However, the HIV-1 Vif protein triggers APOBEC3G degradation, which helps to explain why this innate defense does not protect patients. The APOBEC3G-Vif interaction is a promising therapeutic target, but the benefit of the enabling of HIV-1 restriction in patients is unlikely to be known until Vif antagonists are developed. As a necessary prelude to such studies, cell-based HIV-1 evolution experiments were done to find out whether APOBEC3G can provide a long-term block to Vif-deficient virus replication and, if so, whether HIV-1 variants that resist restriction would emerge. APOBEC3G-expressing T cells were infected with Vif-deficient HIV-1. Virus infectivity was suppressed in 45/48 cultures for more than five weeks, but replication was eventually detected in three cultures. Virus-growth characteristics and sequencing demonstrated that these isolates were still Vif-deficient and that in fact, these viruses had acquired a promoter mutation and a Vpr null mutation. Resistance occurred by a novel tolerance mechanism in which the resistant viruses packaged less APOBEC3G and accumulated fewer hypermutations. These data support the development of antiretrovirals that antagonize Vif and thereby enable endogenous APOBEC3G to suppress HIV-1 replication.
Figures
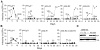

References
-
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. - PubMed
-
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. - PubMed
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
-
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

