The Q-cycle reviewed: How well does a monomeric mechanism of the bc(1) complex account for the function of a dimeric complex?
- PMID: 18501698
- PMCID: PMC2578832
- DOI: 10.1016/j.bbabio.2008.04.037
The Q-cycle reviewed: How well does a monomeric mechanism of the bc(1) complex account for the function of a dimeric complex?
Abstract
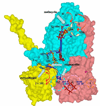
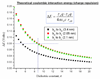
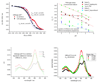
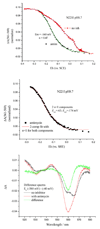
Recent progress in understanding the Q-cycle mechanism of the bc(1) complex is reviewed. The data strongly support a mechanism in which the Q(o)-site operates through a reaction in which the first electron transfer from ubiquinol to the oxidized iron-sulfur protein is the rate-determining step for the overall process. The reaction involves a proton-coupled electron transfer down a hydrogen bond between the ubiquinol and a histidine ligand of the [2Fe-2S] cluster, in which the unfavorable protonic configuration contributes a substantial part of the activation barrier. The reaction is endergonic, and the products are an unstable ubisemiquinone at the Q(o)-site, and the reduced iron-sulfur protein, the extrinsic mobile domain of which is now free to dissociate and move away from the site to deliver an electron to cyt c(1) and liberate the H(+). When oxidation of the semiquinone is prevented, it participates in bypass reactions, including superoxide generation if O(2) is available. When the b-heme chain is available as an acceptor, the semiquinone is oxidized in a process in which the proton is passed to the glutamate of the conserved -PEWY- sequence, and the semiquinone anion passes its electron to heme b(L) to form the product ubiquinone. The rate is rapid compared to the limiting reaction, and would require movement of the semiquinone closer to heme b(L) to enhance the rate constant. The acceptor reactions at the Q(i)-site are still controversial, but likely involve a "two-electron gate" in which a stable semiquinone stores an electron. Possible mechanisms to explain the cyt b(150) phenomenon are discussed, and the information from pulsed-EPR studies about the structure of the intermediate state is reviewed. The mechanism discussed is applicable to a monomeric bc(1) complex. We discuss evidence in the literature that has been interpreted as shown that the dimeric structure participates in a more complicated mechanism involving electron transfer across the dimer interface. We show from myxothiazol titrations and mutational analysis of Tyr-199, which is at the interface between monomers, that no such inter-monomer electron transfer is detected at the level of the b(L) hemes. We show from analysis of strains with mutations at Asn-221 that there are coulombic interactions between the b-hemes in a monomer. The data can also be interpreted as showing similar coulombic interaction across the dimer interface, and we discuss mechanistic implications.
Figures







References
-
- Osyczka A, Moser CC, Dutton PL. Fixing the Q-cycle. Trends in Biochemical Science. 2005;30:176–182. - PubMed
-
- Crofts AR. The cytochrome bc1 complex – function in the context of structure. Annu. Rev. Physiol. 2004;vol. 66:689–733. - PubMed
-
- Crofts AR. The bc1 complex: what is there left to argue about? In: Wikström M, editor. Biophysical and Structural Aspects of Bioenergetics. Cambridge: Royal Society of Chemistry Publishing; 2005. pp. 123–155.
-
- Crofts AR. Proton-coupled electron transfer at the Qo-site of the bc1 complex controls the rate of ubihydroquinone oxidation. Biochim. Biophys. Acta. 2004;1655:77–92. - PubMed
-
- Crofts AR, Lhee S, Crofts SB, Cheng J, Rose S. Proton pumping in the bc1 complex: A new gating mechanism that prevents short circuits. Biochim. Biophys. Acta. 2006;1757:1019–1034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

