Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase
- PMID: 18502750
- PMCID: PMC2459273
- DOI: 10.1074/jbc.M800019200
Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase
Abstract
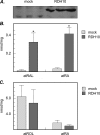
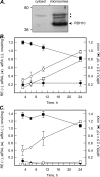
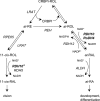
Human retinol dehydrogenase 10 (RDH10) was implicated in the oxidation of all-trans-retinol for biosynthesis of all-trans-retinoic acid, however, initial assays suggested that RDH10 prefers NADP(+) as a cofactor, undermining its role as an oxidative enzyme. Here, we present evidence that RDH10 is, in fact, a strictly NAD(+)-dependent enzyme with multisubstrate specificity that recognizes cis-retinols as well as all-trans-retinol as substrates. RDH10 has a relatively high apparent K(m) value for NAD(+) (~100 microm) but the lowest apparent K(m) value for all-trans-retinol (~0.035 microm) among all NAD(+)-dependent retinoid oxidoreductases. Due to its high affinity for all-trans-retinol, RDH10 exhibits a greater rate of retinol oxidation in the presence of cellular retinol-binding protein type I (CRBPI) than human microsomal RoDH4, but like RoDH4, RDH10 does not recognize retinol bound to CRBPI as a substrate. Consistent with its preference for NAD(+), RDH10 functions exclusively in the oxidative direction in the cells, increasing the levels of retinaldehyde and retinoic acid. Targeted small interfering RNA-mediated silencing of endogenous RDH10 or RoDH4 expression in human cells results in a significant decrease in retinoic acid production from retinol, identifying both human enzymes as physiologically relevant retinol dehydrogenases. The dual cis/trans substrate specificity suggests a dual physiological role for RDH10: in the biosynthesis of 11-cis-retinaldehyde for vision as well as the biosynthesis of all-trans-retinoic acid for differentiation and development.
Figures


References
-
- Mangelsdorf, D., Umesono, K., and Evans, R. M. (1994) in The Retinoids: Biology, Chemistry and Medicine (Sporn, M. B., Roberts, A. B., and Goodman, D. S., eds) pp. 319-350, Raven Press, New York
-
- Napoli, J. L. (1999) Biochim. Biophys. Acta 1440 139-162 - PubMed
-
- Duester, G. (1996) Biochemistry 35 12221-12227 - PubMed
-
- Kedishvili, N. Y. (2002) Curr. Org. Chem. 6 1247-1257
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

