Protein Z-dependent protease inhibitor binds to the C-terminal domain of protein Z
- PMID: 18502758
- PMCID: PMC2459281
- DOI: 10.1074/jbc.M802639200
Protein Z-dependent protease inhibitor binds to the C-terminal domain of protein Z
Abstract
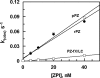
Protein Z (PZ) is a multidomain vitamin K-dependent plasma protein that functions as a cofactor to promote the inactivation of factor Xa (fXa) by PZ-dependent protease inhibitor (ZPI) by three orders of magnitude. To understand the mechanism by which PZ improves the reactivity of fXa with ZPI, we expressed wild-type PZ, PZ lacking the gamma-carboxyglutamic acid domain (GD-PZ), and a chimeric PZ mutant in which both Gla and EGF-like domains of the molecule were substituted with identical domains of fXa. The ZPI binding and the cofactor function of the PZ derivatives were characterized in both binding and kinetic assays. The binding assay indicated that all PZ derivatives interact with ZPI with a similar dissociation constant (K(D)) of approximately 7 nm. However, the apparent K(D) for the chimeric PZ-mediated ZPI inhibition of fXa was elevated 6-fold on PC/PS vesicles and its capacity to function as a cofactor to accelerate the ZPI inhibition of fXa was also decreased 6-fold. The cofactor activity of GD-PZ was dramatically impaired; however, the deletion mutant exhibited a normal cofactor function in solution. A chimeric activated protein C mutant containing the Gla domain of fXa was susceptible to inhibition by ZPI in the presence of PZ. These results suggest that: (i) the ZPI interactive site of PZ is located within the C-terminal domain of the cofactor and (ii) a specific interaction between the Gla domains of PZ and fXa contributes approximately 6-fold to the acceleration of the ZPI inhibition of fXa on phospholipid membranes.
Figures



References
-
- Ichinose, A., Takeya, H., Espling, E., Iwanaga, S., Kisiel, W., and Davie, E. W. (1990) Biochem. Biophys. Res. Commun. 172 1139–1144 - PubMed
-
- Sejima, H., Hayashi, T., Deyashiki, Y., Nishioka, J., and Suzuki, K. (1990) Biochem. Biophys. Res. Commun. 171 661–668 - PubMed
-
- Han, X., Fiehler, R., and Broze, G. J., Jr. (2000) Blood 96 3049–3055 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

