Rad50 is not essential for the Mre11-dependent repair of DNA double-strand breaks in Halobacterium sp. strain NRC-1
- PMID: 18502851
- PMCID: PMC2493270
- DOI: 10.1128/JB.00292-08
Rad50 is not essential for the Mre11-dependent repair of DNA double-strand breaks in Halobacterium sp. strain NRC-1
Abstract
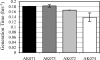
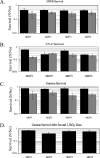
The genome of the halophilic archaeon Halobacterium sp. strain NRC-1 encodes homologs of the eukaryotic Mre11 and Rad50 proteins, which are involved in the recognition and end processing of DNA double-strand breaks in the homologous recombination repair pathway. We have analyzed the phenotype of Halobacterium deletion mutants lacking mre11 and/or rad50 after exposure to UV-C radiation, an alkylating agent (N-methyl-N'-nitro-N-nitrosoguanidine), and gamma radiation, none of which resulted in a decrease in survival of the mutant strains compared to that of the background strain. However, a decreased rate of repair of DNA double-strand breaks in strains lacking the mre11 gene was observed using pulsed-field gel electrophoresis. These observations led to the hypothesis that Mre11 is essential for the repair of DNA double-strand breaks in Halobacterium, whereas Rad50 is dispensable. This is the first identification of a Rad50-independent function for the Mre11 protein, and it represents a shift in the Archaea away from the eukaryotic model of homologous recombination repair of DNA double-strand breaks.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

