Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation
- PMID: 18503043
- PMCID: PMC2423659
- DOI: 10.1093/jxb/ern114
Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation
Abstract
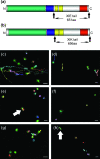
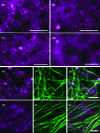
Although organelle movement in higher plants is predominantly actin-based, potential roles for the 17 predicted Arabidopsis myosins in motility are only just emerging. It is shown here that two Arabidopsis myosins from class XI, XIE, and XIK, are involved in Golgi, peroxisome, and mitochondrial movement. Expression of dominant negative forms of the myosin lacking the actin binding domain at the amino terminus perturb organelle motility, but do not completely inhibit movement. Latrunculin B, an actin destabilizing drug, inhibits organelle movement to a greater extent compared to the effects of AtXIE-T/XIK-T expression. Amino terminal YFP fusions to XIE-T and XIK-T are dispersed throughout the cytosol and do not completely decorate the organelles whose motility they affect. XIE-T and XIK-T do not affect the global actin architecture, but their movement and location is actin-dependent. The potential role of these truncated myosins as genetically encoded inhibitors of organelle movement is discussed.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

