Substrate cleavage analysis of furin and related proprotein convertases. A comparative study
- PMID: 18505722
- PMCID: PMC2475709
- DOI: 10.1074/jbc.M803762200
Substrate cleavage analysis of furin and related proprotein convertases. A comparative study
Abstract
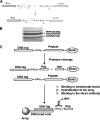
We present the data and the technology, a combination of which allows us to determine the identity of proprotein convertases (PCs) related to the processing of specific protein targets including viral and bacterial pathogens. Our results, which support and extend the data of other laboratories, are required for the design of effective inhibitors of PCs because, in general, an inhibitor design starts with a specific substrate. Seven proteinases of the human PC family cleave the multibasic motifs R-X-(R/K/X)-R downward arrow and, as a result, transform proproteins, including those from pathogens, into biologically active proteins and peptides. The precise cleavage preferences of PCs have not been known in sufficient detail; hence we were unable to determine the relative importance of the individual PCs in infectious diseases, thus making the design of specific inhibitors exceedingly difficult. To determine the cleavage preferences of PCs in more detail, we evaluated the relative efficiency of furin, PC2, PC4, PC5/6, PC7, and PACE4 in cleaving over 100 decapeptide sequences representing the R-X-(R/K/X)-R downward arrow motifs of human, bacterial, and viral proteins. Our computer analysis of the data and the follow-on cleavage analysis of the selected full-length proteins corroborated our initial results thus allowing us to determine the cleavage preferences of the PCs and to suggest which PCs are promising drug targets in infectious diseases. Our results also suggest that pathogens, including anthrax PA83 and the avian influenza A H5N1 (bird flu) hemagglutinin precursor, evolved to be as sensitive to PC proteolysis as the most sensitive normal human proteins.
Figures


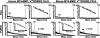



References
-
- Seidah, N. G., Day, R., and Chretien, M. (1993) Biochem. Soc. Trans. 21685 -691 - PubMed
-
- Steiner, D. F. (1998) Curr. Opin. Chem. Biol. 231 -39 - PubMed
-
- Fugere, M., Limperis, P. C., Beaulieu-Audy, V., Gagnon, F., Lavigne, P., Klarskov, K., Leduc, R., and Day, R. (2002) J. Biol. Chem. 2777648 -7656 - PubMed
-
- Taylor, N. A., Van De Ven, W. J., and Creemers, J. W. (2003) FASEB J. 171215 -1227 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

