Bayesian LASSO for quantitative trait loci mapping
- PMID: 18505874
- PMCID: PMC2429858
- DOI: 10.1534/genetics.107.085589
Bayesian LASSO for quantitative trait loci mapping
Abstract
The mapping of quantitative trait loci (QTL) is to identify molecular markers or genomic loci that influence the variation of complex traits. The problem is complicated by the facts that QTL data usually contain a large number of markers across the entire genome and most of them have little or no effect on the phenotype. In this article, we propose several Bayesian hierarchical models for mapping multiple QTL that simultaneously fit and estimate all possible genetic effects associated with all markers. The proposed models use prior distributions for the genetic effects that are scale mixtures of normal distributions with mean zero and variances distributed to give each effect a high probability of being near zero. We consider two types of priors for the variances, exponential and scaled inverse-chi(2) distributions, which result in a Bayesian version of the popular least absolute shrinkage and selection operator (LASSO) model and the well-known Student's t model, respectively. Unlike most applications where fixed values are preset for hyperparameters in the priors, we treat all hyperparameters as unknowns and estimate them along with other parameters. Markov chain Monte Carlo (MCMC) algorithms are developed to simulate the parameters from the posteriors. The methods are illustrated using well-known barley data.
Figures

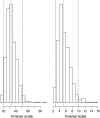
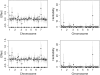



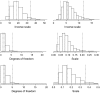
References
-
- Andrews, D. F., and C. L. Mallows, 1974. Scale mixtures of normal distributions. J. R. Stat. Soc. Ser. B 36 99–102.
-
- Bao, K., and B. K. Mallick, 2004. Gene selection using a two-level hierarchical Bayesian model. Bioinformatics 20 3423–3430. - PubMed
-
- Chhikara, R. S., and L. Folks, 1989. The Inverse Gaussian Distribution: Theory, Methodology, and Applications. Marcel Dekker, New York.
-
- Efron, B., T. Hastie, I. Johnstone and R. Tibshirani, 2004. Least angle regression. Ann. Stat. 32 407–499.
-
- Figueiredo, M. A. T., 2003. Adaptive sparseness for supervised learning. IEEE Trans. Patt. Anal. Machine Intell. 25 1150–1159.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

