Neutral sphingomyelinase-3 is a DNA damage and nongenotoxic stress-regulated gene that is deregulated in human malignancies
- PMID: 18505924
- PMCID: PMC2642592
- DOI: 10.1158/1541-7786.MCR-07-2097
Neutral sphingomyelinase-3 is a DNA damage and nongenotoxic stress-regulated gene that is deregulated in human malignancies
Abstract
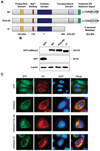
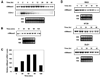
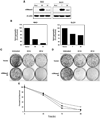
In this study, we report the characterization of a novel genotoxic and nongenotoxic stress-regulated gene that we had previously named as SKNY. Our results indicate that SKNY encodes the recently identified neutral sphingomyelinase-3 (nSMase3; hereafter SKNY is referred to as nSMase3). Examination of nSMase3 subcellular distribution reveals nSMase3 to localize to the endoplasmic reticulum (ER), and deletion of a COOH-terminal region containing its putative transmembrane domain and ER targeting signal partly alters its compartmentalization to the ER. Treatment with genotoxic Adriamycin and nongenotoxic tumor necrosis factor-alpha up-regulates endogenous nSMase3 expression, albeit with different kinetics. Tumor necrosis factor-alpha up-regulates nSMase3 expression within 2 h that lasts beyond 24 h and declines to control levels by 36 h. Adriamycin up-regulation of nSMase3 is transient, occurs within 30 min, and declines to control levels by 120 min. Prolonged treatment with Adriamycin by 24 h and beyond, however, causes a down-regulation in nSMase3 expression. Activation of wild-type p53 also down-regulates nSMase3 expression, suggesting that DNA damage-mediated nSMase3 down-regulation seems to occur partly through the tumor suppressor p53. Overexpression of exogenous nSMase3 sensitizes cells to Adriamycin-induced cell killing, a finding consistent with the proposed proapoptotic role of nSMase enzymes and nSMase-generated ceramide. We further investigated nSMase3 expression in various human malignancies and found its expression to be deregulated in several types of primary tumors when compared with their matching normal tissues. Collectively, our results have identified nSMase3 to be an important molecule that is linked to tumorigenesis and cellular stress response.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures



References
-
- Zheng W, Kollmeyer J, Symolon H, et al. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. - PubMed
-
- Gulbins E, Li PL. Physiological and pathophysiological aspects of ceramide. Am J Physiol Regul Integr Comp Physiol. 2006;290:R11–R26. - PubMed
-
- Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

