Predicting helix orientation for coiled-coil dimers
- PMID: 18506779
- PMCID: PMC2562949
- DOI: 10.1002/prot.22118
Predicting helix orientation for coiled-coil dimers
Abstract
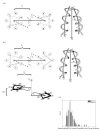
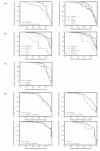
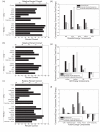
The alpha-helical coiled coil is a structurally simple protein oligomerization or interaction motif consisting of two or more alpha helices twisted into a supercoiled bundle. Coiled coils can differ in their stoichiometry, helix orientation, and axial alignment. Because of the near degeneracy of many of these variants, coiled coils pose a challenge to fold recognition methods for structure prediction. Whereas distinctions between some protein folds can be discriminated on the basis of hydrophobic/polar patterning or secondary structure propensities, the sequence differences that encode important details of coiled-coil structure can be subtle. This is emblematic of a larger problem in the field of protein structure and interaction prediction: that of establishing specificity between closely similar structures. We tested the behavior of different computational models on the problem of recognizing the correct orientation--parallel vs. antiparallel--of pairs of alpha helices that can form a dimeric coiled coil. For each of 131 examples of known structure, we constructed a large number of both parallel and antiparallel structural models and used these to assess the ability of five energy functions to recognize the correct fold. We also developed and tested three sequence-based approaches that make use of varying degrees of implicit structural information. The best structural methods performed similarly to the best sequence methods, correctly categorizing approximately 81% of dimers. Steric compatibility with the fold was important for some coiled coils we investigated. For many examples, the correct orientation was determined by smaller energy differences between parallel and antiparallel structures distributed over many residues and energy components. Prediction methods that used structure but incorporated varying approximations and assumptions showed quite different behaviors when used to investigate energetic contributions to orientation preference. Sequence based methods were sensitive to the choice of residue-pair interactions scored.
Figures



References
-
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18(4):617–625. - PubMed
-
- McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22(3):356–358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

