Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer
- PMID: 18506891
- PMCID: PMC2762280
- DOI: 10.1002/hep.22283
Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer
Abstract
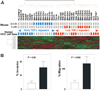
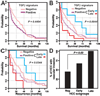
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world. The clinical heterogeneity of HCC, and the lack of good diagnostic markers and treatment strategies, has rendered the disease a major challenge. Patients with HCC have a highly variable clinical course, indicating that HCC comprises several biologically distinctive subgroups reflecting a molecular heterogeneity of the tumors. Transforming growth factor beta (TGF-beta) is known to exhibit tumor stage dependent suppressive (that is, growth inhibition) and oncogenic (that is, invasiveness) properties. Here, we asked if a TGF-beta specific gene expression signature could refine the classification and prognostic predictions for HCC patients. Applying a comparative functional genomics approach we demonstrated that a temporal TGF-beta gene expression signature established in mouse primary hepatocytes successfully discriminated distinct subgroups of HCC. The TGF-beta positive cluster included two novel homogeneous groups of HCC associated with early and late TGF-beta signatures. Kaplan-Meier plots and log-rank statistics indicated that the patients with a late TGF-beta signature showed significantly (P < 0.005) shortened mean survival time (16.2 +/- 5.3 months) compared to the patients with an early (60.7 +/- 16.1 months) TGF-beta signature. Also, tumors expressing late TGF-beta-responsive genes displayed invasive phenotype and increased tumor recurrence. We also showed that the late TGF-beta signature accurately predicted liver metastasis and discriminated HCC cell lines by degree of invasiveness. Finally, we established that the TGF-beta gene expression signature possessed a predictive value for tumors other than HCC.
Conclusion: These data demonstrate the clinical significance of the genes embedded in TGF-beta expression signature for the molecular classification of HCC.
Conflict of interest statement
Potential conflict of interest: Nothing to report.
Figures



References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- El Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127(5 Suppl 1):S27–S34. - PubMed
-
- Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. - PubMed
-
- Bruix J, Boix L, Sala M, Llovet JM. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. - PubMed
-
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
