Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation
- PMID: 18508253
- PMCID: PMC2504688
- DOI: 10.1016/j.ceb.2008.03.019
Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation
Abstract
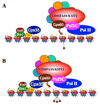
Chromosomal surfaces are ornamented with a variety of post-translational modifications of histones, which are required for the regulation of many of the DNA-templated processes. Such histone modifications include acetylation, sumoylation, phosphorylation, ubiquitination, and methylation. Histone modifications can either function by disrupting chromosomal contacts or by regulating non-histone protein interactions with chromatin. In this review, recent findings will be discussed regarding the regulation of the implementation and physiological significance for one such histone modification, histone H3 lysine 4 (H3K4) methylation by the yeast COMPASS and mammalian COMPASS-like complexes.
Figures


References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. - PubMed
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

