The role of programmed-1 ribosomal frameshifting in coronavirus propagation
- PMID: 18508552
- PMCID: PMC2435135
- DOI: 10.2741/3046
The role of programmed-1 ribosomal frameshifting in coronavirus propagation
Abstract
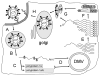
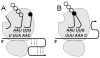
Coronaviruses have the potential to cause significant economic, agricultural and health problems. The severe acute respiratory syndrome (SARS) associated coronavirus outbreak in late 2002, early 2003 called attention to the potential damage that coronaviruses could cause in the human population. The ensuing research has enlightened many to the molecular biology of coronaviruses. A programmed -1 ribosomal frameshift is required by coronaviruses for the production of the RNA dependent RNA polymerase which in turn is essential for viral replication. The frameshifting signal encoded in the viral genome has additional features that are not essential for frameshifting. Elucidation of the differences between coronavirus frameshift signals and signals from other viruses may help our understanding of these features. Here we summarize current knowledge and add additional insight regarding the function of the programmed -1 ribosomal frameshift signal in the coronavirus lifecycle.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
