Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax
- PMID: 18508761
- PMCID: PMC2475714
- DOI: 10.1074/jbc.M800375200
Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax
Abstract
During animal development, distinct tissues, organs, and appendages are specified through differential gene transcription by Hox transcription factors. However, the conserved Hox homeodomains bind DNA with high affinity yet low specificity. We have therefore explored the structure of the Drosophila melanogaster Hox protein Ultrabithorax and the impact of its nonhomeodomain regions on DNA binding properties. Computational and experimental approaches identified several conserved, intrinsically disordered regions outside the homeodomain of Ultrabithorax that impact DNA binding by the homeodomain. Full-length Ultrabithorax bound to target DNA 2.5-fold weaker than its isolated homeodomain. Using N-terminal and C-terminal deletion mutants, we demonstrate that the YPWM region and the disordered microexons (termed the I1 region) inhibit DNA binding approximately 2-fold, whereas the disordered I2 region inhibits homeodomain-DNA interaction a further approximately 40-fold. Binding is restored almost to homeodomain affinity by the mostly disordered N-terminal 174 amino acids (R region) in a length-dependent manner. Both the I2 and R regions contain portions of the activation domain, functionally linking DNA binding and transcription regulation. Given that (i) the I1 region and a portion of the R region alter homeodomain-DNA binding as a function of pH and (ii) an internal deletion within I1 increases Ultrabithorax-DNA affinity, I1 must directly impact homeodomain-DNA interaction energetics. However, I2 appears to indirectly affect DNA binding in a manner countered by the N terminus. The amino acid sequences of I2 and much of the I1 and R regions vary significantly among Ultrabithorax orthologues, potentially diversifying Hox-DNA interactions.
Figures

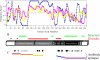
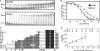
 ) and UbxHD (▪). Each point
in a curve was derived from three replicates within a single experiment, and
the error bars indicate the S.D. for these replicates. For clarity,
only the fraction of free DNA is shown. C, sequence schematic for
UbxIa and its variants. The homeodomain (HD) and activation domain,
which is subdivided into the core region required for function (AC)
and the enhancement region that boosts activity (AER), and a partial
repression domain (R)
(31,
75,
76) are indicated. The highly
conserved YPWM (Exd interaction) motif (Y), five moderately conserved
motifs (1–5), the polyglycine region (G), and the
microexons (mI and mII) are also indicated by bars
below. The sequence boundaries for the 11 deletion mutants and UbxHD
relative to these domains are also shown. D, note the significant
differences in binding affinity. Each value represents at least nine
Kd measurements at pH 7.5. The low affinities of N139 and
N174 are near the experimental limits of gel retardation assays and therefore
have larger errors. Data for UbxIa and the first five N-terminal deletion
mutants fit to a line with R2 = 0.92, indicating that
affinity is linearly dependent on sequence length for these regions.
) and UbxHD (▪). Each point
in a curve was derived from three replicates within a single experiment, and
the error bars indicate the S.D. for these replicates. For clarity,
only the fraction of free DNA is shown. C, sequence schematic for
UbxIa and its variants. The homeodomain (HD) and activation domain,
which is subdivided into the core region required for function (AC)
and the enhancement region that boosts activity (AER), and a partial
repression domain (R)
(31,
75,
76) are indicated. The highly
conserved YPWM (Exd interaction) motif (Y), five moderately conserved
motifs (1–5), the polyglycine region (G), and the
microexons (mI and mII) are also indicated by bars
below. The sequence boundaries for the 11 deletion mutants and UbxHD
relative to these domains are also shown. D, note the significant
differences in binding affinity. Each value represents at least nine
Kd measurements at pH 7.5. The low affinities of N139 and
N174 are near the experimental limits of gel retardation assays and therefore
have larger errors. Data for UbxIa and the first five N-terminal deletion
mutants fit to a line with R2 = 0.92, indicating that
affinity is linearly dependent on sequence length for these regions.

 ) to 40AB was measured over the
pH range 5.0–10.0 in Tris binding buffer at constant ionic strength.
Results are similar to measurements in a phosphate-based buffer from 5.0 to
8.0. B and G depict the binding curves of UbxHD, UbxHK,
N235, N139, N88, and UbxIa, respectively. The binding affinities of UbxIa and
these critical variants were measured at pH 7.5 (▪) and pH 6.0
(
) to 40AB was measured over the
pH range 5.0–10.0 in Tris binding buffer at constant ionic strength.
Results are similar to measurements in a phosphate-based buffer from 5.0 to
8.0. B and G depict the binding curves of UbxHD, UbxHK,
N235, N139, N88, and UbxIa, respectively. The binding affinities of UbxIa and
these critical variants were measured at pH 7.5 (▪) and pH 6.0
( ) in Tris binding buffer.
Binding curves were derived from replicates within a single experiment, which
includes three individual affinity measurements, and the error bars
indicate the S.D. for these replicates. Data from multiple experiments for
each mutant are summarized in Table
1.
) in Tris binding buffer.
Binding curves were derived from replicates within a single experiment, which
includes three individual affinity measurements, and the error bars
indicate the S.D. for these replicates. Data from multiple experiments for
each mutant are summarized in Table
1.


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

