GPR56 regulates pial basement membrane integrity and cortical lamination
- PMID: 18509043
- PMCID: PMC2504715
- DOI: 10.1523/JNEUROSCI.0853-08.2008
GPR56 regulates pial basement membrane integrity and cortical lamination
Abstract
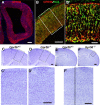
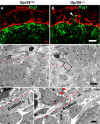
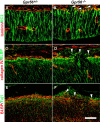
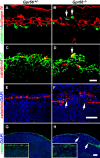
GPR56 is a member of the family of adhesion G-protein-coupled receptors that have a large extracellular region containing a GPS (G-protein proteolytic site) domain. Loss-of-function mutations in the GPR56 gene cause a specific human brain malformation called bilateral frontoparietal polymicrogyria (BFPP). BFPP is a radiological diagnosis and its histopathology remains unclear. This study demonstrates that loss of the mouse Gpr56 gene leads to neuronal ectopia in the cerebral cortex, a cobblestone-like cortical malformation. There are four crucial events in the development of cobblestone cortex, namely defective pial basement membrane (BM), abnormal anchorage of radial glial endfeet, mislocalized Cajal-Retzius cells, and neuronal overmigration. By detailed time course analysis, we reveal that the leading causal events are likely the breaches in the pial BM. We show further that GPR56 is present in abundance in radial glial endfeet. Furthermore, a putative ligand of GPR56 is localized in the marginal zone or overlying extracellular matrix. These observations provide compelling evidence that GPR56 functions in regulating pial BM integrity during cortical development.
Figures



References
-
- Anton ES, Kreidberg JA, Rakic P. Distinct functions of alpha3 and alpha (v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 1999;22:277–289. - PubMed
-
- Chang BS, Piao X, Bodell A, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, Grant PE, Barkovich AJ, Walsh CA. Bilateral frontoparietal polymicrogyria: clinical and radiological features in 10 families with linkage to chromosome 16. Ann Neurol. 2003;53:596–606. - PubMed
-
- De Arcangelis A, Mark M, Kreidberg J, Sorokin L, Georges-Labouesse E. Synergistic activities of alpha3 and alpha6 integrins are required during apical ectodermal ridge formation and organogenesis in the mouse. Development. 1999;126:3957–3968. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
