Pharmacogenetic pathway analysis of docetaxel elimination
- PMID: 18509327
- PMCID: PMC4612590
- DOI: 10.1038/clpt.2008.95
Pharmacogenetic pathway analysis of docetaxel elimination
Abstract
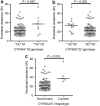
The purpose of this study was to evaluate the affinity of docetaxel for 14 transporter proteins and assess the functional significance of 17 variants in five genes involved in drug elimination. Among the transfected models investigated, OATP1B3 (SLCO1B3) was identified as the most efficient influx transporter for docetaxel. None of the observed genotypes (SLCO1B3, ABCB1, and ABCC2) was related with docetaxel clearance in 92 white patients (P > 0.17). However, the simultaneous presence of the CYP3A4*1B and CYP3A5*1A alleles was associated with a 64% increase in docetaxel clearance (P = 0.0015), independent of both sex and CYP3A activity (as determined using the erythromycin breath test). This haplotype was also associated with increased midazolam clearance in another population (P = 0.0198). An analysis of the CYP3A locus among CEPH-HapMap samples revealed that CYP3A4*1B is present exclusively among a subset of CYP3A5 expressors. Therefore, future studies should first stratify the population on the basis of CYP3A5 genotype and then compare CYP3A activity between individuals with and without the CYP3A4*1B allele.
Conflict of interest statement
Figures
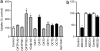
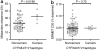


References
-
- Baker SD, Sparreboom A, Verweij J. Clinical pharmacokinetics of docetaxel: recent developments. Clin Pharmacokinet. 2006;45:235–252. - PubMed
-
- Bruno R, et al. Population pharmacokinetics/pharmacodynamics of docetaxel in phase II studies in patients with cancer. J Clin Oncol. 1998;16:187–196. - PubMed
-
- Bruno R, et al. Alpha-1-acid glycoprotein as an independent predictor for treatment effects and a prognostic factor of survival in patients with non-small cell lung cancer treated with docetaxel. Clin Cancer Res. 2003;9:1077–1082. - PubMed
-
- Veyrat-Follet C, Bruno R, Olivares R, Rhodes GR, Chaikin P. Clinical trial simulation of docetaxel in patients with cancer as a tool for dosage optimization. Clin Pharmacol Ther. 2000;68:677–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

