Evolution of the CDKN1C-KCNQ1 imprinted domain
- PMID: 18510768
- PMCID: PMC2427030
- DOI: 10.1186/1471-2148-8-163
Evolution of the CDKN1C-KCNQ1 imprinted domain
Abstract
Background: Genomic imprinting occurs in both marsupial and eutherian mammals. The CDKN1C and IGF2 genes are both imprinted and syntenic in the mouse and human, but in marsupials only IGF2 is imprinted. This study examines the evolution of features that, in eutherians, regulate CDKN1C imprinting.
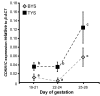
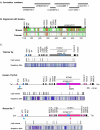
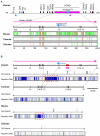
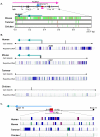
Results: Despite the absence of imprinting, CDKN1C protein was present in the tammar wallaby placenta. Genomic analysis of the tammar region confirmed that CDKN1C is syntenic with IGF2. However, there are fewer LTR and DNA elements in the region and in intron 9 of KCNQ1. In addition there are fewer LINEs in the tammar compared with human and mouse. While the CpG island in intron 10 of KCNQ1 and promoter elements could not be detected, the antisense transcript KCNQ1OT1 that regulates CDKN1C imprinting in human and mouse is still expressed.
Conclusion: CDKN1C has a conserved function, likely antagonistic to IGF2, in the mammalian placenta that preceded its acquisition of imprinting. CDKN1C resides in synteny with IGF2, demonstrating that imprinting of the two genes did not occur concurrently to balance maternal and paternal influences on the growth of the placenta. The expression of KCNQ1OT1 in the absence of CDKN1C imprinting suggests that antisense transcription at this locus preceded imprinting of this domain. These findings demonstrate the stepwise accumulation of control mechanisms within imprinted domains and show that CDKN1C imprinting cannot be due to its synteny with IGF2 or with its placental expression in mammals.
Figures


References
-
- Killian JK, Nolan CM, Stewart N. et al.Monotreme IGF2 expression and ancestral origin of genomic imprinting. J ExpZool. 2001;291(2):205–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

