Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin
- PMID: 18511462
- PMCID: PMC2475631
- DOI: 10.1093/nar/gkn340
Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin
Abstract
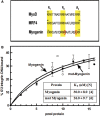
Four myogenic regulatory factors (MRFs); MyoD, Myf-5, MRF4 and Myogenin direct muscle tissue differentiation. Heterodimers of MRFs with E-proteins activate muscle-specific gene expression by binding to E-box motifs d(CANNTG) in their promoters or enhancers. We showed previously that in contrast to the favored binding of E-box by MyoD-E47 heterodimers, homodimeric MyoD associated preferentially with quadruplex structures of regulatory sequences of muscle-specific genes. To inquire whether other MRFs shared the DNA binding preferences of MyoD, the DNA affinities of hetero- and homo-dimeric MyoD, MRF4 and Myogenin were compared. Similarly to MyoD, heterodimers with E47 of MRF4 or Myogenin bound E-box more tightly than quadruplex DNA. However, unlike homodimeric MyoD or MRF4, Myogenin homodimers associated weakly and nonpreferentially with quadruplex DNA. By reciprocally switching basic regions between MyoD and Myogenin we demonstrated dominance of MyoD in determining the quadruplex DNA-binding affinity. Thus, Myogenin with an implanted MyoD basic region bound quadruplex DNA nearly as tightly as MyoD. However, a grafted Myogenin basic region did not diminish the high affinity of homodimeric MyoD for quadruplex DNA. We speculate that the dissimilar interaction of MyoD and Myogenin with tetrahelical domains in muscle gene promoters may differently regulate their myogenic activities.
Figures

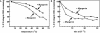


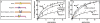
Similar articles
-
Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes.J Biol Chem. 2005 Jul 22;280(29):26805-12. doi: 10.1074/jbc.M500820200. Epub 2005 May 27. J Biol Chem. 2005. PMID: 15923190
-
Analysis of the DNA-binding properties of MyoD, myogenin, and E12 by fluorescence anisotropy.Biochemistry. 2002 Sep 3;41(35):10888-94. doi: 10.1021/bi025528q. Biochemistry. 2002. PMID: 12196028
-
Differential interactions of Id proteins with basic-helix-loop-helix transcription factors.J Biol Chem. 1997 Aug 8;272(32):19785-93. doi: 10.1074/jbc.272.32.19785. J Biol Chem. 1997. PMID: 9242638
-
Regulation and functions of myogenic regulatory factors in lower vertebrates.Comp Biochem Physiol B Biochem Mol Biol. 2001 Aug;130(1):1-12. doi: 10.1016/s1096-4959(01)00412-2. Comp Biochem Physiol B Biochem Mol Biol. 2001. PMID: 11470439 Review.
-
Determination versus differentiation and the MyoD family of transcription factors.Biochem Cell Biol. 1995 Sep-Oct;73(9-10):723-32. doi: 10.1139/o95-080. Biochem Cell Biol. 1995. PMID: 8714693 Review.
Cited by
-
The miR-6240 target gene Igf2bp3 promotes myoblast fusion by enhancing myomaker mRNA stability.Cell Mol Biol Lett. 2024 Dec 5;29(1):152. doi: 10.1186/s11658-024-00650-1. Cell Mol Biol Lett. 2024. PMID: 39639214 Free PMC article.
-
Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells.Nucleic Acids Res. 2009 Jul;37(13):4194-204. doi: 10.1093/nar/gkn1076. Epub 2009 Feb 11. Nucleic Acids Res. 2009. PMID: 19211664 Free PMC article.
-
Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide.Nucleic Acids Res. 2011 Oct;39(18):8005-16. doi: 10.1093/nar/gkr536. Epub 2011 Jul 4. Nucleic Acids Res. 2011. PMID: 21729868 Free PMC article.
-
Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation.Nucleic Acids Res. 2013 Jan 7;41(1):76-89. doi: 10.1093/nar/gks1071. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161683 Free PMC article.
-
Mechanisms of Binding Specificity among bHLH Transcription Factors.Int J Mol Sci. 2021 Aug 24;22(17):9150. doi: 10.3390/ijms22179150. Int J Mol Sci. 2021. PMID: 34502060 Free PMC article. Review.
References
-
- Tajbakhsh S. Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Curr. Opin. Genet. Dev. 2003;13:413–422. - PubMed
-
- Schnorrer F, Dickson BJ. Muscle building; mechanisms of myotube guidance and attachment site selection. Dev. Cell. 2004;7:9–20. - PubMed
-
- Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. - PubMed
-
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. - PubMed
-
- Munsterberg AE, Kitajewski J, Bumcrot DA, McMahon AP, Lassar AB. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9:2911–2922. - PubMed

