Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer
- PMID: 18511480
- PMCID: PMC2556025
- DOI: 10.1074/mcp.M800029-MCP200
Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer
Abstract
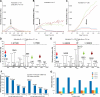
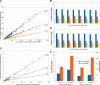
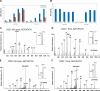
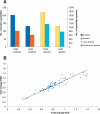
Isobaric stable isotope tagging reagents such as tandem mass tags or isobaric tags for relative and absolute quantification enable multiplexed quantification of peptides via reporter ion signals in the low mass range of tandem mass spectra. Until recently, the poor recovery of low mass fragments observed in tandem mass spectra acquired on ion trap mass spectrometers precluded the use of these reagents on this widely available instrument platform. The Pulsed Q Dissociation (PQD) technique allows negotiating this limitation but suffers from poor fragmentation efficiency, which has raised doubts in the community as to its practical utility. Here we show that by carefully optimizing instrument parameters such as collision energy, activation Q, delay time, ion isolation width, number of microscans, and number of trapped ions, low m/z fragment ion intensities can be generated that enable accurate peptide quantification at the 100 amol level. Side by side comparison of PQD on an LTQ Orbitrap with CID on a five-year old Q-Tof Ultima using complex protein digests shows that whereas precision of quantification of 10-15% can be achieved by both approaches, PQD quantifies twice as many proteins. PQD on an LTQ Orbitrap also outperforms "higher energy collision induced dissociation" on the same instrument using the recently introduced octapole collision cell in terms of lower limit of quantification. Finally, we demonstrate the significant analytical potential of iTRAQ quantification using PQD on an LTQ Orbitrap by quantitatively measuring the kinase interaction profile of the small molecule drug imatinib in K-562 cells. This article gives practical guidance for the implementation of PQD, discusses its merits, and for the first time, compares its performance to higher energy collision-induced dissociation.
Figures

References
-
- Bantscheff, M., Schirle, M., Sweetman, G., Rick, J., and Kuster, B. ( 2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 - PubMed
-
- Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 - PubMed
-
- Wang, Y. K., Ma, Z., Quinn, D. F., and Fu, E. W. ( 2001) Inverse 18O labeling mass spectrometry for the rapid identification of marker/target proteins. Anal. Chem. 73, 3742–3750 - PubMed
-
- Wang, Y. K., Ma, Z., Quinn, D. F., and Fu, E. W. ( 2002) Inverse 15N-metabolic labeling/mass spectrometry for comparative proteomics and rapid identification of protein markers/targets. Rapid Commun. Mass Spectrom. 16, 1389–1397 - PubMed
-
- Wang, Y. K., Quinn, D. F., Ma, Z., and Fu, E. W. ( 2002) Inverse labeling-mass spectrometry for the rapid identification of differentially expressed protein markers/targets. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 782, 291–306 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

