Functional and structural characterization of DR_0079 from Deinococcus radiodurans, a novel Nudix hydrolase with a preference for cytosine (deoxy)ribonucleoside 5'-Di- and triphosphates
- PMID: 18512963
- PMCID: PMC2867059
- DOI: 10.1021/bi800099d
Functional and structural characterization of DR_0079 from Deinococcus radiodurans, a novel Nudix hydrolase with a preference for cytosine (deoxy)ribonucleoside 5'-Di- and triphosphates
Abstract
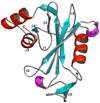
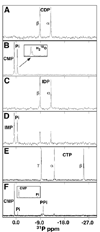
The genome of the extremely radiation resistant bacterium Deinococcus radiodurans encodes 21 Nudix hydrolases, of which only two have been characterized in detail. Here we report the activity and crystal structure for DR_0079, the first Nudix hydrolase observed to have a marked preference for cytosine ribonucleoside 5'-diphosphate (CDP) and cytosine ribonucleoside 5'-triphosphate (CTP). After CDP and CTP, the next most preferred substrates for DR_0079, with a relative activity of <50%, were the corresponding deoxyribose nucleotides, dCDP and dCTP. Hydrolase activity at the site of the phosphodiester bond was corroborated using (31)P NMR spectroscopy to follow the phosphorus resonances for three substrates, CDP, IDP, and CTP, and their hydrolysis products, CMP + P(i), IMP + P(i), and CMP + PP(i), respectively. Nucleophilic substitution at the beta-phosphorus of CDP and CTP was established, using (31)P NMR spectroscopy, by the appearance of an upfield-shifted P(i) resonance and line-broadened PP(i) resonance, respectively, when the hydrolysis was performed in 40% H(2)(18)O-enriched water. The optimal activity for CDP was at pH 9.0-9.5 with the reaction requiring divalent metal cation (Mg(2+) > Mn(2+) > Co(2+)). The biochemical data are discussed with reference to the crystal structure for DR_0079 that was determined in the metal-free form at 1.9 A resolution. The protein contains nine beta-strands, three alpha-helices, and two 3(10)-helices organized into three subdomains: an N-terminal beta-sheet, a central Nudix core, and a C-terminal helix-turn-helix motif. As observed for all known structures of Nudix hydrolases, the alpha-helix of the "Nudix box" is one of two helices that sandwich a "four-strand" mixed beta-sheet. To identify residues potentially involved in metal and substrate binding, NMR chemical shift mapping experiments were performed on (15)N-labeled DR_0079 with the paramagnetic divalent cation Co(2+) and the nonhydrolyzable substrate thymidine 5'-O-(alpha,beta-methylenediphosphate) and the results mapped onto the crystal structure.
Figures

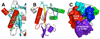
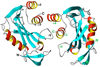


Similar articles
-
Solution structure of hypothetical Nudix hydrolase DR0079 from extremely radiation-resistant Deinococcus radiodurans bacterium.Proteins. 2004 Jul 1;56(1):28-39. doi: 10.1002/prot.20082. Proteins. 2004. PMID: 15162484
-
Characterization of a nudix hydrolase from Deinococcus radiodurans with a marked specificity for (deoxy)ribonucleoside 5'-diphosphates.BMC Biochem. 2004 May 17;5:7. doi: 10.1186/1471-2091-5-7. BMC Biochem. 2004. PMID: 15147580 Free PMC article.
-
Structure of an N-terminally truncated Nudix hydrolase DR2204 from Deinococcus radiodurans.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Nov 1;65(Pt 11):1083-7. doi: 10.1107/S1744309109037191. Epub 2009 Oct 13. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19923723 Free PMC article.
-
Structures and mechanisms of Nudix hydrolases.Arch Biochem Biophys. 2005 Jan 1;433(1):129-43. doi: 10.1016/j.abb.2004.08.017. Arch Biochem Biophys. 2005. PMID: 15581572 Review.
-
Solution structure and mechanism of the MutT pyrophosphohydrolase.Adv Enzymol Relat Areas Mol Biol. 1999;73:183-207. doi: 10.1002/9780470123195.ch6. Adv Enzymol Relat Areas Mol Biol. 1999. PMID: 10218109 Review.
Cited by
-
Antifungal symbiotic peptide NCR044 exhibits unique structure and multifaceted mechanisms of action that confer plant protection.Proc Natl Acad Sci U S A. 2020 Jul 7;117(27):16043-16054. doi: 10.1073/pnas.2003526117. Epub 2020 Jun 22. Proc Natl Acad Sci U S A. 2020. PMID: 32571919 Free PMC article.
-
The evolution of function within the Nudix homology clan.Proteins. 2017 May;85(5):775-811. doi: 10.1002/prot.25223. Epub 2017 Mar 16. Proteins. 2017. PMID: 27936487 Free PMC article.
-
Structure of a Nudix hydrolase (MutT) in the Mg(2+)-bound state from Bartonella henselae, the bacterium responsible for cat scratch fever.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Sep 1;67(Pt 9):1078-83. doi: 10.1107/S1744309111011559. Epub 2011 Aug 16. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21904053 Free PMC article.
-
Substrate specificity characterization for eight putative nudix hydrolases. Evaluation of criteria for substrate identification within the Nudix family.Proteins. 2016 Dec;84(12):1810-1822. doi: 10.1002/prot.25163. Epub 2016 Oct 1. Proteins. 2016. PMID: 27618147 Free PMC article.
-
The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications.AMB Express. 2019 Sep 3;9(1):138. doi: 10.1186/s13568-019-0862-x. AMB Express. 2019. PMID: 31482336 Free PMC article. Review.
References
-
- Abeygunawardana C, Weber DJ, Gittis AG, Frick DN, Lin J, Miller AF, Bessman MJ, Mildvan AS. Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry. 1995;34:14997–15005. - PubMed
-
- Bessman MJ, Frick N, O’Handley SF. The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J. Biol. Chem. 1996;271:25059–25062. - PubMed
-
- Dunn CA, O’Handley SF, Frick DN, Bessman MJ. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J. Biol. Chem. 1999;274:32318–32324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

