CUSP: an algorithm to distinguish structurally conserved and unconserved regions in protein domain alignments and its application in the study of large length variations
- PMID: 18513436
- PMCID: PMC2423364
- DOI: 10.1186/1472-6807-8-28
CUSP: an algorithm to distinguish structurally conserved and unconserved regions in protein domain alignments and its application in the study of large length variations
Abstract
Background: Distantly related proteins adopt and retain similar structural scaffolds despite length variations that could be as much as two-fold in some protein superfamilies. In this paper, we describe an analysis of indel regions that accommodate length variations amongst related proteins. We have developed an algorithm CUSP, to examine multi-membered PASS2 superfamily alignments to identify indel regions in an automated manner. Further, we have used the method to characterize the length, structural type and biochemical features of indels in related protein domains.
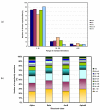
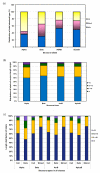
Results: CUSP, examines protein domain structural alignments to distinguish regions of conserved structure common to related proteins from structurally unconserved regions that vary in length and type of structure. On a non-redundant dataset of 353 domain superfamily alignments from PASS2, we find that 'length- deviant' protein superfamilies show > 30% length variation from their average domain length. 60% of additional lengths that occur in indels are short-length structures (< 5 residues) while 6% of indels are > 15 residues in length. Structural types in indels also show class-specific trends.
Conclusion: The extent of length variation varies across different superfamilies and indels show class-specific trends for preferred lengths and structural types. Such indels of different lengths even within a single protein domain superfamily could have structural and functional consequences that drive their selection, underlying their importance in similarity detection and computational modelling. The availability of systematic algorithms, like CUSP, should enable decision making in a domain superfamily-specific manner.
Figures


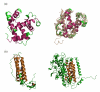

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

