The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering
- PMID: 18515383
- PMCID: PMC2527263
- DOI: 10.1529/biophysj.107.122465
The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering
Abstract
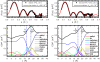
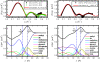
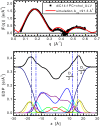
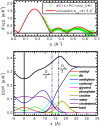
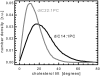
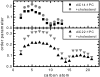
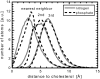
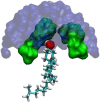
We investigate the structure of cholesterol-containing membranes composed of either short-chain (diC14:1PC) or long-chain (diC22:1PC) monounsaturated phospholipids. Bilayer structural information is derived from all-atom molecular dynamics simulations, which are validated via direct comparison to x-ray scattering experiments. We show that the addition of 40 mol % cholesterol results in a nearly identical increase in the thickness of the two different bilayers. In both cases, the chain ordering dominates over the hydrophobic matching between the length of the cholesterol molecule and the hydrocarbon thickness of the bilayer, which one would expect to cause a thinning of the diC22:1PC bilayer. For both bilayers there is substantial headgroup rearrangement for lipids directly in contact with cholesterol, supporting the so-called umbrella model. Importantly, in diC14:1PC bilayers, a dynamic network of hydrogen bonds stabilizes long-lived reorientations of some cholesterol molecules, during which they are found to lie perpendicular to the bilayer normal, deep within the bilayer's hydrophobic core. Additionally, the simulations show that the diC14:1PC bilayer is significantly more permeable to water. These differences may be correlated with faster cholesterol flip-flop between the leaflets of short-chain lipid bilayers, resulting in an asymmetric distribution of cholesterol molecules. This asymmetry was observed experimentally in a case of unilamellar vesicles (ULVs), and reproduced through a set of novel asymmetric simulations. In contrast to ULVs, experimental data for oriented multilamellar stacks does not show the asymmetry, suggesting that it results from the curvature of the ULV bilayers.
Figures



References
-
- Lee, A. G. 2004. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 1666:62–87. - PubMed
-
- Cornelius, F., N. Turner, and H. R. Christensen. 2003. Modulation of Na,K-ATPase by phospholipids and cholesterol. II. Steady-state and presteady-state kinetics. Biochemistry. 42:8541–8549. - PubMed
-
- Bretscher, M. S., and S. Munro. 1993. Cholesterol and the Golgi apparatus. Science. 261:1280–1281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

